- Importing the libraries, and merge dataset
- Analyse the features, Feature selection.
- Preprocessing and Cleaning.
- Train and Test split Dataset
- Models training and hyper parameter tunning
- Conclude model with better results.
The training files can be found in the training_files branch
https://github.com/Emekaborisama/disease_symptom_classification/tree/training_files
The API code can be found in the master branch
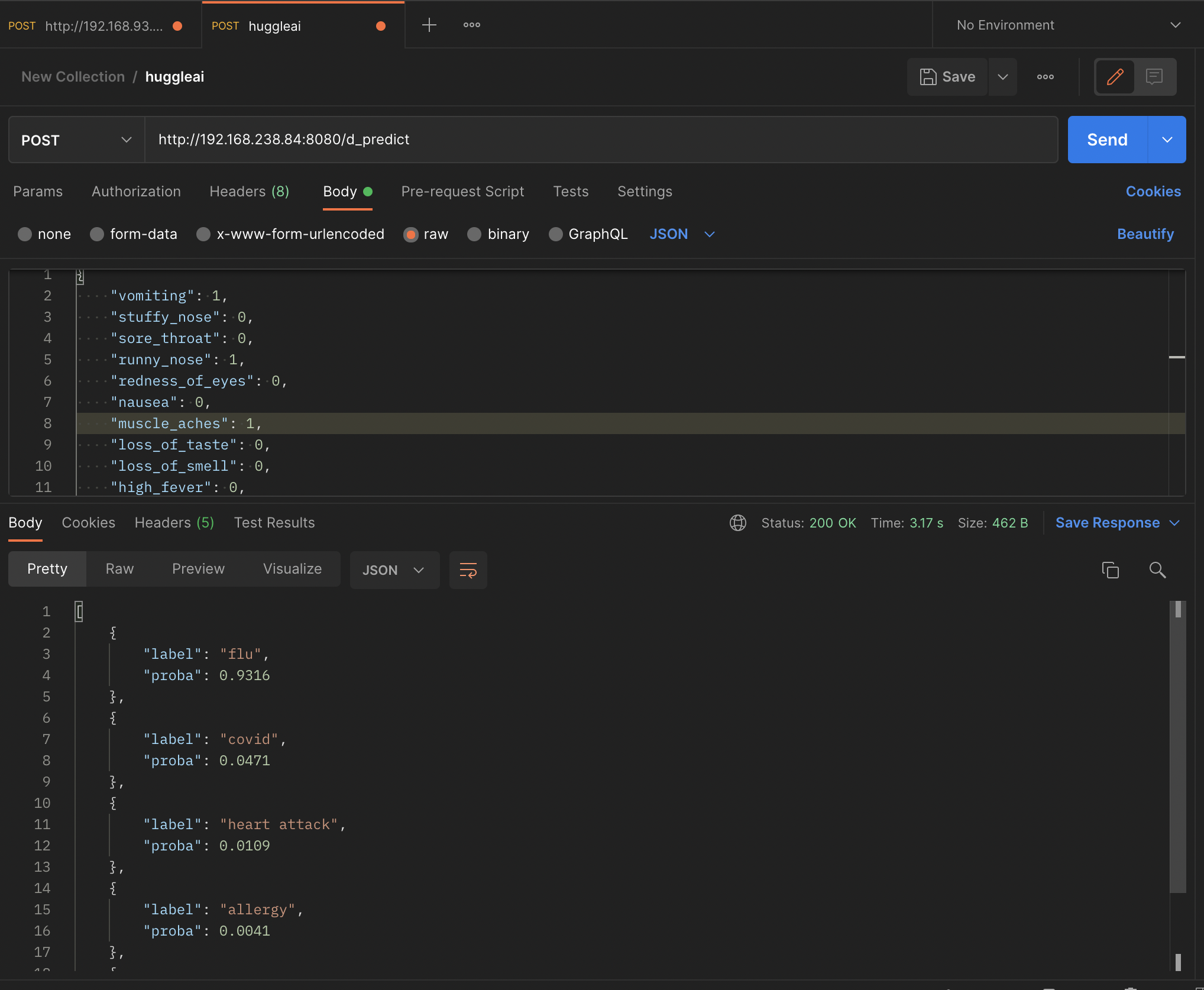
Conclusion: In all we have been able to build a baseline model that take in MRS. Sarah symptoms and predict the disease she is likely having.
Our model with Grandient boosting has 92% accuracy (F1 Score) and we solely don't want to depend on that accuracy that why our future work will require us to deliberately and intentionally test our model to ensure we don't give the 'Never Google symptoms prediction'.
Future work:
- Hyper paramter tunning
- Adding more dataset
- Detecting and handling imbalance features
- Plot confusion metrics
- Bias and variance trade off
- Behaviour testing
- Deploying to Kubernetes pod or Docker