Keras implementation of DeepSEA, original version published in Nature Methods 2015 here.
- May 6, 2020: wrap into a package for re-use (especially on reading data)
After cloned the Github repository, initialize a new
Anaconda environment using the conda_env.yml
configurations.
First download the compiled dataset from DeepSEA official site here: http://deepsea.princeton.edu/help/
Put the download *.mat files under ./data/ folder.
For the trained parameters file, download from sourceforge: https://sourceforge.net/projects/bionas/files/DeepSEA_v0.9/
Now there are two different routes to go:
- if loading from the original torch parameters: download
deepsea_cpu.pthtodatafolder, then open python terminal:
from load_torch_weights import build_and_load_weights
model = build_and_load_weights()Note: This file is a torch7 file with all trained parameters in an OrderedDict python object, when loaded with
torch.loadmethod.
The t7 file can be downloaded here
- if loading from the converted Keras model, download the Keras parameter file:
from model import load_model
model = load_model()The keras h5 file can be downloaded here.
Make sure we did not do anything crazy; the results should be the same regardless of which model building route you took:
from evaluate import val_performance
import numpy as np
val_evals = val_performance(model)
print(np.nanmean(val_evals['auroc'])) # 0.937
print(np.nanmean(val_evals['aupr'])) # 0.402Also we can evaluate the test-data performance. This might take a bit longer using a single GPU, so we can convert the Keras model to a Multi-GPU model:
Note: Remember to toggle the
gpusargument to the available gpus you have.
from evaluate import test_performance
from model import convert_to_multigpu
mg_model = convert_to_multigpu(model, gpus=3)
test_evals = test_performance(mg_model)
print(np.nanmean(test_evals['auroc'])) # 0.931
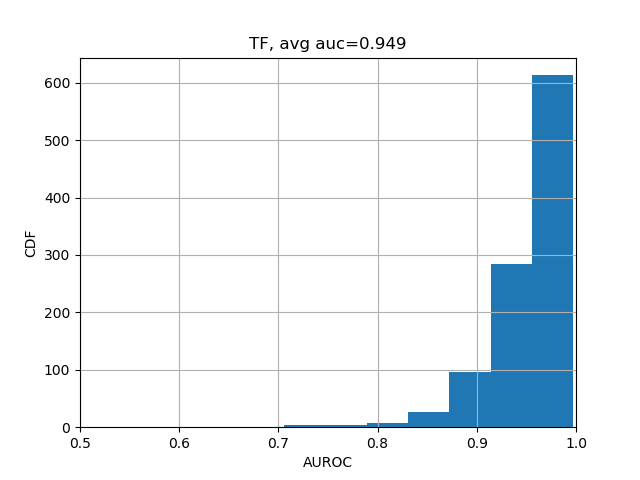
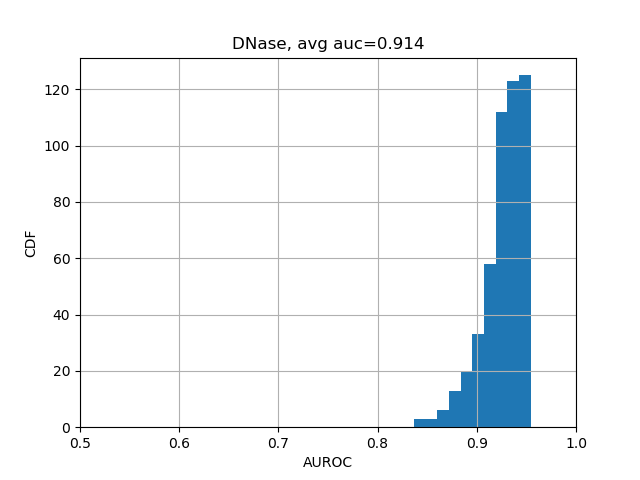
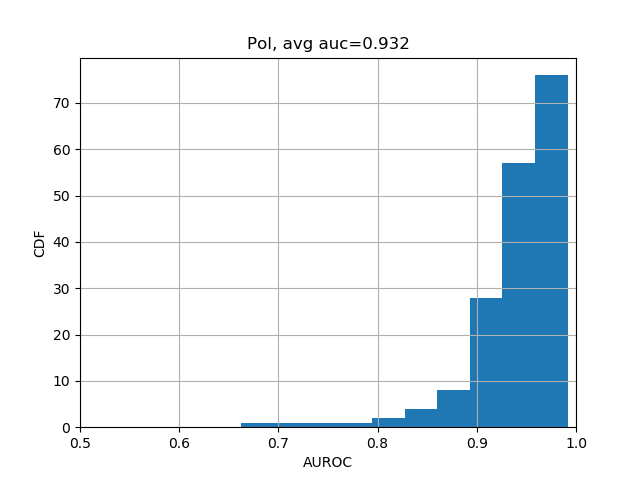
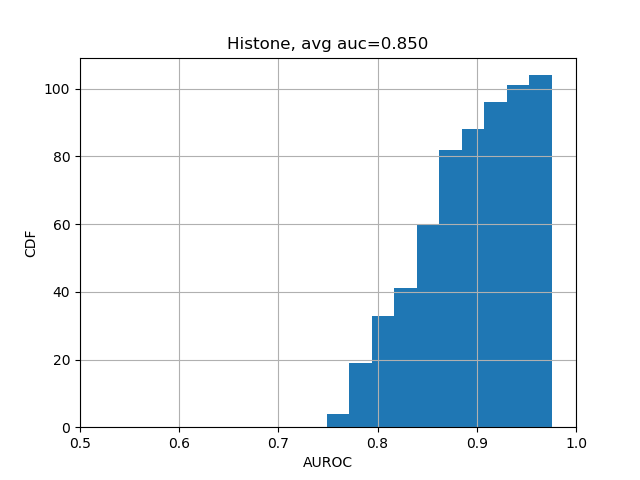
print(np.nanmean(test_evals['aupr'])) # 0.338The results should be close enough to "average AUC of 0.933 and AUPRC of 0.342", as stated in the Selene biorxiv manuscript at the bottom of Page 5.
We can further break down the prediction accuracies by their categories:
from plot import plot_auc
plot_auc(test_evals)The results should look like the following