2017-05-02
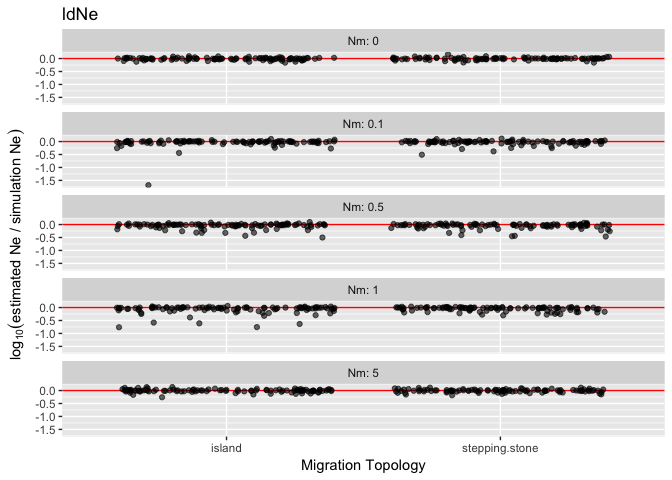
The data was generated with a fastsimcoal simulation that created allele frequencies at equilibrium and were then used to initialize and run 5 generations of an rmetasim simulation.
The scenarios were composed of the following parameter combinations:
scenario Ne Nm theta mig.type num.loci num.pops div.time
1 1 50 0.0 0.2 island 1000 5 25000
2 2 500 0.0 0.2 island 1000 5 25000
3 3 50 0.1 0.2 island 1000 5 25000
4 4 500 0.1 0.2 island 1000 5 25000
5 5 50 0.5 0.2 island 1000 5 25000
6 6 500 0.5 0.2 island 1000 5 25000
7 7 50 1.0 0.2 island 1000 5 25000
8 8 500 1.0 0.2 island 1000 5 25000
9 9 50 5.0 0.2 island 1000 5 25000
10 10 500 5.0 0.2 island 1000 5 25000
11 11 50 0.0 0.2 stepping.stone 1000 5 25000
12 12 500 0.0 0.2 stepping.stone 1000 5 25000
13 13 50 0.1 0.2 stepping.stone 1000 5 25000
14 14 500 0.1 0.2 stepping.stone 1000 5 25000
15 15 50 0.5 0.2 stepping.stone 1000 5 25000
16 16 500 0.5 0.2 stepping.stone 1000 5 25000
17 17 50 1.0 0.2 stepping.stone 1000 5 25000
18 18 500 1.0 0.2 stepping.stone 1000 5 25000
19 19 50 5.0 0.2 stepping.stone 1000 5 25000
20 20 500 5.0 0.2 stepping.stone 1000 5 25000
mut.rate mig.rate
1 1e-03 0e+00
2 1e-04 0e+00
3 1e-03 2e-03
4 1e-04 2e-04
5 1e-03 1e-02
6 1e-04 1e-03
7 1e-03 2e-02
8 1e-04 2e-03
9 1e-03 1e-01
10 1e-04 1e-02
11 1e-03 0e+00
12 1e-04 0e+00
13 1e-03 2e-03
14 1e-04 2e-04
15 1e-03 1e-02
16 1e-04 1e-03
17 1e-03 2e-02
18 1e-04 2e-03
19 1e-03 1e-01
20 1e-04 1e-02
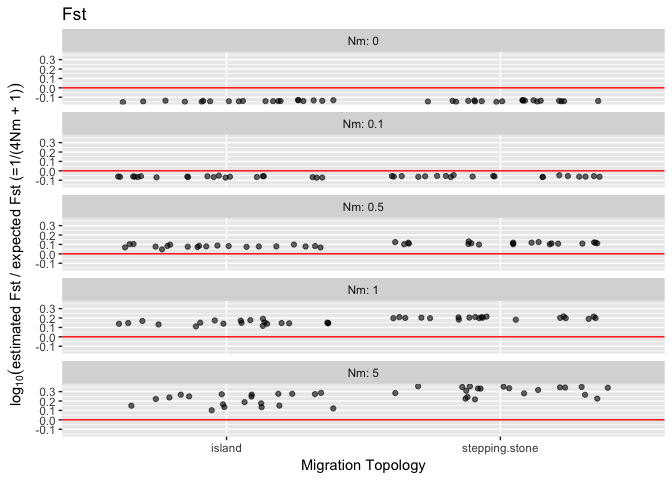
The "island" model specifies a migration matrix such as the following from scenario 3, where the migration rate for a population is 0.002 split among the other 4 populations:
[,1] [,2] [,3] [,4] [,5]
[1,] 0.9980 0.0005 0.0005 0.0005 0.0005
[2,] 0.0005 0.9980 0.0005 0.0005 0.0005
[3,] 0.0005 0.0005 0.9980 0.0005 0.0005
[4,] 0.0005 0.0005 0.0005 0.9980 0.0005
[5,] 0.0005 0.0005 0.0005 0.0005 0.9980
The "stepping.stone" model specifies a migration matrix such as the following from scenario 15, where the migration rate for a population is 0.01 split between the neighboring two populations:
[,1] [,2] [,3] [,4] [,5]
[1,] 0.990 0.005 0.000 0.000 0.005
[2,] 0.005 0.990 0.005 0.000 0.000
[3,] 0.000 0.005 0.990 0.005 0.000
[4,] 0.000 0.000 0.005 0.990 0.005
[5,] 0.005 0.000 0.000 0.005 0.990
All output files are contained in the folder ".sim.data", where "" defaults to "sim.results.YYYMMDD.HHMM". Each scenario has gtypes objects stored in a R workspace file, named "gtypes.sc.rdata" where "sc" is the scenario number. This file contains two objects:
fsc.list- A list of gtypes from fastsimcoal, one per replicate. The scenario parameters are stored as a one row data.frame inattr(fsc.list, "scenario").rms.list- A list of gtypes from rmetasim after initialization with the corresponding gtypes object fromfsc.list. This contains the final genotypes. The scenario parameters are also stored as a one row data.frame inattr(rms.list, "scenario").
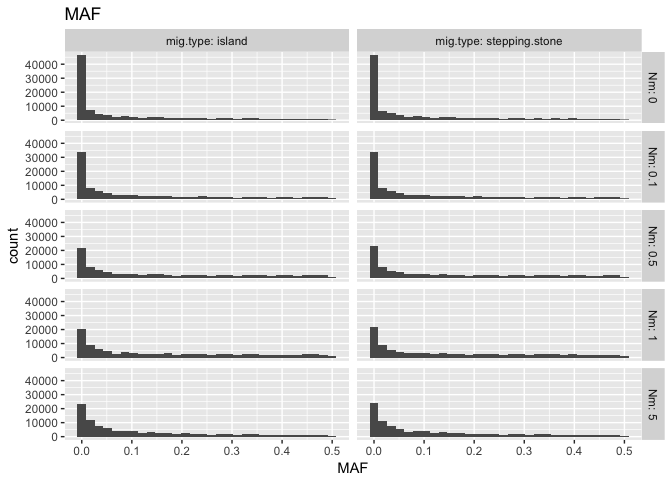
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.