Like juicer, but without all those calories
dietJuicer is a lighter-weight, HPC flexible version of juicer written with snakemake.
dietJuicerCore is designed to be run on a single Hi-C library. Sequencing replicates (i.e. the same Hi-C library split to maximize complexity), can combined and run together. Run the following processing steps for each Hi-C library:
-
Clone workflow into working directory:
git clone https://github.com/EricSDavis/dietJuicer.git . -
Edit the tab-separated
samplesheet.txtfile with the names ofRead1andRead2gzipped fastq files and the path to these files under theSequencing_Directorycolumn. No naming convention is needed for fastq files, as long as they are gzipped. At minimum one additional column is required to determine which combination of files to combine into a group (see step 3). Optional columns can be specified below to capture sample metadata, as shown below:Project Cell_Type Genotype Bio_Rep Tech_Rep Seq_Rep Read1 Read2 Sequencing_Directory PROJ CELL WT 1 1 1 sample1_R1.fq.gz sample1_R2.fq.gz /path/to/fastq/directory/ PROJ CELL WT 2 1 1 sample2_R1.fq.gz sample2_R2.fq.gz /path/to/fastq/directory/ PROJ CELL WT 3 1 1 sample3_R1.fq.gz sample3_R2.fq.gz /path/to/fastq/directory/ PROJ CELL WT 4 1 1 sample4_R1.fq.gz sample4_R2.fq.gz /path/to/fastq/directory/ PROJ CELL MUT 1 1 1 sample5_R1.fq.gz sample5_R2.fq.gz /path/to/fastq/directory/ PROJ CELL MUT 2 1 1 sample6_R1.fq.gz sample6_R2.fq.gz /path/to/fastq/directory/ PROJ CELL MUT 3 1 1 sample7_R1.fq.gz sample7_R2.fq.gz /path/to/fastq/directory/ PROJ CELL MUT 4 1 1 sample8_R1.fq.gz sample8_R2.fq.gz /path/to/fastq/directory/ PROJ CELL WT 1 1 2 sample9_R1.fq.gz sample9_R2.fq.gz /path/to/fastq/directory/ PROJ CELL WT 2 1 2 sample10_R1.fq.gz sample10_R2.fq.gz /path/to/fastq/directory/ PROJ CELL WT 3 1 2 sample11_R1.fq.gz sample11_R2.fq.gz /path/to/fastq/directory/ PROJ CELL WT 4 1 2 sample12_R1.fq.gz sample12_R2.fq.gz /path/to/fastq/directory/ PROJ CELL MUT 1 1 2 sample13_R1.fq.gz sample13_R2.fq.gz /path/to/fastq/directory/ PROJ CELL MUT 2 1 2 sample14_R1.fq.gz sample14_R2.fq.gz /path/to/fastq/directory/ PROJ CELL MUT 3 1 2 sample15_R1.fq.gz sample15_R2.fq.gz /path/to/fastq/directory/ PROJ CELL MUT 4 1 2 sample16_R1.fq.gz sample16_R2.fq.gz /path/to/fastq/directory/ -
Edit
config/config.yamlfile for your system/experiment. ThegroupByparameter uses a list of column names to join as the group id. This parameter minimally requires one column and will generate a group name prefix before all output files. See the example below:## Path to sample sheet samplesheet: 'samplesheet.txt' ## List columns to group (i.e. any columns left out will become part of the same group) groupBy: ['Project', 'Cell_Type', 'Genotype', 'Time', 'Bio_Rep', 'Tech_Rep'] ## Genome-specific reference parameters fasta: '/proj/seq/data/HG19_UCSC/Sequence/BWAIndex/genome.fa' chromSizes: '/proj/phanstiel_lab/software/resources/hg19_chromSizes.txt' ## Restriction site information site: "MboI" # or 'none' site_file: 'restriction_sites/hg19_MboI_chr.txt' # Make sure chromosomes match bwa index chromosomes (i.e. both start with "chr") ligation: "GATCGATC" ## Set splitsize splitsize: 200000000 ## 200000000 good default (50M reads/file) (i.e. 4000000 = 1M reads/file) ## Java memory for buildHiC (hic/norm rules) javaMem: "250880" ## Additional options mapq0_reads_included: 0
-
Submit to
SLURMwith sbatch script:sbatch dietJuicerCore.sh
dietJuicerCore.shwill submit a long-running, low-resource job script that will spawn other jobs in the pipeline as dependencies are fulfilled. For systems other than slurm, editdietJuicerCore.shto create a submission script for your HPC's job scheduler.
After running these steps the pipeline will produce the following files:
output/{group}/{group}_dedup_merged_nodups.txtoutput/{group}/{group}_inter.txtoutput/{group}/{group}_inter_30.txtoutput/{group}/{group}_inter.hicoutput/{group}/{group}_inter_30.hic
Output directory structure:
slurm-{jobid}.out
output/
├── logs_slurm
└── {group}
├── benchmarks
│ ├── {group}_{rule}_split{splitName}.tsv
│ └── ...
├── logs
│ ├── {group}_{rule}_split{splitName}.err
│ └── ...
├── {group}_dedup_merged_nodups.txt.gz
├── {group}_inter_30.hic
├── {group}_inter_30_hists.m
├── {group}_inter_30.txt
├── {group}_inter.hic
├── {group}_inter_hists.m
├── {group}_inter.txt
├── {group}_splitR1_done.txt
├── {group}_splitR2_done.txt
├── splitsR1
│ ├── {splitName}_R1.fastq.gz -- removed
│ └── ...
└── splitsR2
├── {splitName}_R2.fastq.gz -- removed
└── ...
These files can be used as input for creating combined .hic maps (see "Creating a Hi-C Map").
It is recommended to fork this repo and make adjustments for your HPC environment for future Hi-C processing.
A Hi-C map can be created by running dietJuicerCore (see "Quickstart") or by combining the output of several processed libraries to create a "mega" map using dietJuicerMerge. Run the following processing steps to create a combined Hi-C map:
-
Clone workflow into working directory:
git clone https://github.com/EricSDavis/dietJuicer.git . -
Edit the tab-separated
samplesheet.txtfile with the paths tomerged_nodups,inter, andinter30text files. Just likedietJuicerCorea minimum of one additional column is required to determine which combination of files to combine into a group (see step 3). Optional columns can be specified below to capture sample metadata, as shown below:Project Cell_Type Genotype Bio_Rep Tech_Rep Seq_Rep Read1 Read2 Sequencing_Directory merged_nodups inter inter30 PROJ CELL WT 1 1 1 sample1_R1.fq.gz sample1_R2.fq.gz /path/to/fastq/directory/ /path/to/merged_nodups.txt.gz /path/to/inter.txt /path/to/inter_30.txt PROJ CELL WT 2 1 1 sample2_R1.fq.gz sample2_R2.fq.gz /path/to/fastq/directory/ /path/to/merged_nodups.txt.gz /path/to/inter.txt /path/to/inter_30.txt PROJ CELL WT 3 1 1 sample3_R1.fq.gz sample3_R2.fq.gz /path/to/fastq/directory/ /path/to/merged_nodups.txt.gz /path/to/inter.txt /path/to/inter_30.txt PROJ CELL WT 4 1 1 sample4_R1.fq.gz sample4_R2.fq.gz /path/to/fastq/directory/ /path/to/merged_nodups.txt.gz /path/to/inter.txt /path/to/inter_30.txt PROJ CELL MUT 1 1 1 sample5_R1.fq.gz sample5_R2.fq.gz /path/to/fastq/directory/ /path/to/merged_nodups.txt.gz /path/to/inter.txt /path/to/inter_30.txt PROJ CELL MUT 2 1 1 sample6_R1.fq.gz sample6_R2.fq.gz /path/to/fastq/directory/ /path/to/merged_nodups.txt.gz /path/to/inter.txt /path/to/inter_30.txt PROJ CELL MUT 3 1 1 sample7_R1.fq.gz sample7_R2.fq.gz /path/to/fastq/directory/ /path/to/merged_nodups.txt.gz /path/to/inter.txt /path/to/inter_30.txt PROJ CELL MUT 4 1 1 sample8_R1.fq.gz sample8_R2.fq.gz /path/to/fastq/directory/ /path/to/merged_nodups.txt.gz /path/to/inter.txt /path/to/inter_30.txt PROJ CELL WT 1 1 2 sample9_R1.fq.gz sample9_R2.fq.gz /path/to/fastq/directory/ /path/to/merged_nodups.txt.gz /path/to/inter.txt /path/to/inter_30.txt PROJ CELL WT 2 1 2 sample10_R1.fq.gz sample10_R2.fq.gz /path/to/fastq/directory/ /path/to/merged_nodups.txt.gz /path/to/inter.txt /path/to/inter_30.txt PROJ CELL WT 3 1 2 sample11_R1.fq.gz sample11_R2.fq.gz /path/to/fastq/directory/ /path/to/merged_nodups.txt.gz /path/to/inter.txt /path/to/inter_30.txt PROJ CELL WT 4 1 2 sample12_R1.fq.gz sample12_R2.fq.gz /path/to/fastq/directory/ /path/to/merged_nodups.txt.gz /path/to/inter.txt /path/to/inter_30.txt PROJ CELL MUT 1 1 2 sample13_R1.fq.gz sample13_R2.fq.gz /path/to/fastq/directory/ /path/to/merged_nodups.txt.gz /path/to/inter.txt /path/to/inter_30.txt PROJ CELL MUT 2 1 2 sample14_R1.fq.gz sample14_R2.fq.gz /path/to/fastq/directory/ /path/to/merged_nodups.txt.gz /path/to/inter.txt /path/to/inter_30.txt PROJ CELL MUT 3 1 2 sample15_R1.fq.gz sample15_R2.fq.gz /path/to/fastq/directory/ /path/to/merged_nodups.txt.gz /path/to/inter.txt /path/to/inter_30.txt PROJ CELL MUT 4 1 2 sample16_R1.fq.gz sample16_R2.fq.gz /path/to/fastq/directory/ /path/to/merged_nodups.txt.gz /path/to/inter.txt /path/to/inter_30.txt -
Edit
config/config.yamlfile for your system/experiment. ThegroupByparameter uses a list of column names to join as the group id. This parameter minimally requires one column and will generate a group name prefix before all output files. In the example below we combine a mega Hi-C map forWTandMUT(output group names will bePROJ_CELL_WTandPROJ_CELL_MUT, respectively):## Path to sample sheet samplesheet: 'samplesheet.txt' ## List columns to group (i.e. any columns left out will become part of the same group) groupBy: ['Project', 'Cell_Type', 'Genotype'] ## Genome-specific reference parameters fasta: '/proj/seq/data/HG19_UCSC/Sequence/BWAIndex/genome.fa' chromSizes: '/proj/phanstiel_lab/software/resources/hg19_chromSizes.txt' ## Restriction site information site: "MboI" # or 'none' site_file: 'restriction_sites/hg19_MboI_chr.txt' # Make sure chromosomes match bwa index chromosomes (i.e. both start with "chr") ligation: "GATCGATC" ## Set splitsize splitsize: 200000000 ## 200000000 good default (50M reads/file) (i.e. 4000000 = 1M reads/file) ## Java memory for buildHiC (hic/norm rules) javaMem: "250880" ## Additional options mapq0_reads_included: 0
-
Submit to
SLURMwith sbatch script:sbatch dietJuicerMerge.sh
dietJuicerMerge.shwill submit a long-running, low-resource job script that will spawn other jobs in the pipeline as dependencies are fulfilled. For systems other than slurm, editdietJuicerMerge.shto create a submission script for your HPC's job scheduler.
After running these steps the pipeline will produce the following files:
output/{group}/{group}_merged_nodups.txtoutput/{group}/{group}_inter.txtoutput/{group}/{group}_inter_30.txtoutput/{group}/{group}_inter.hicoutput/{group}/{group}_inter_30.hic
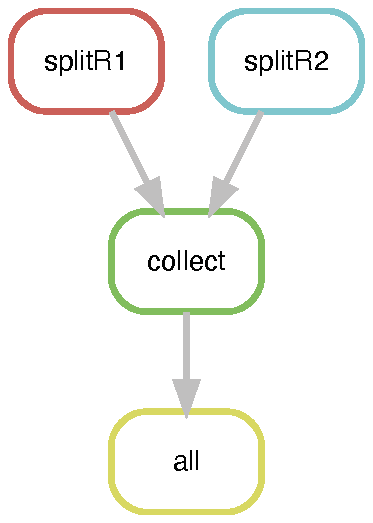
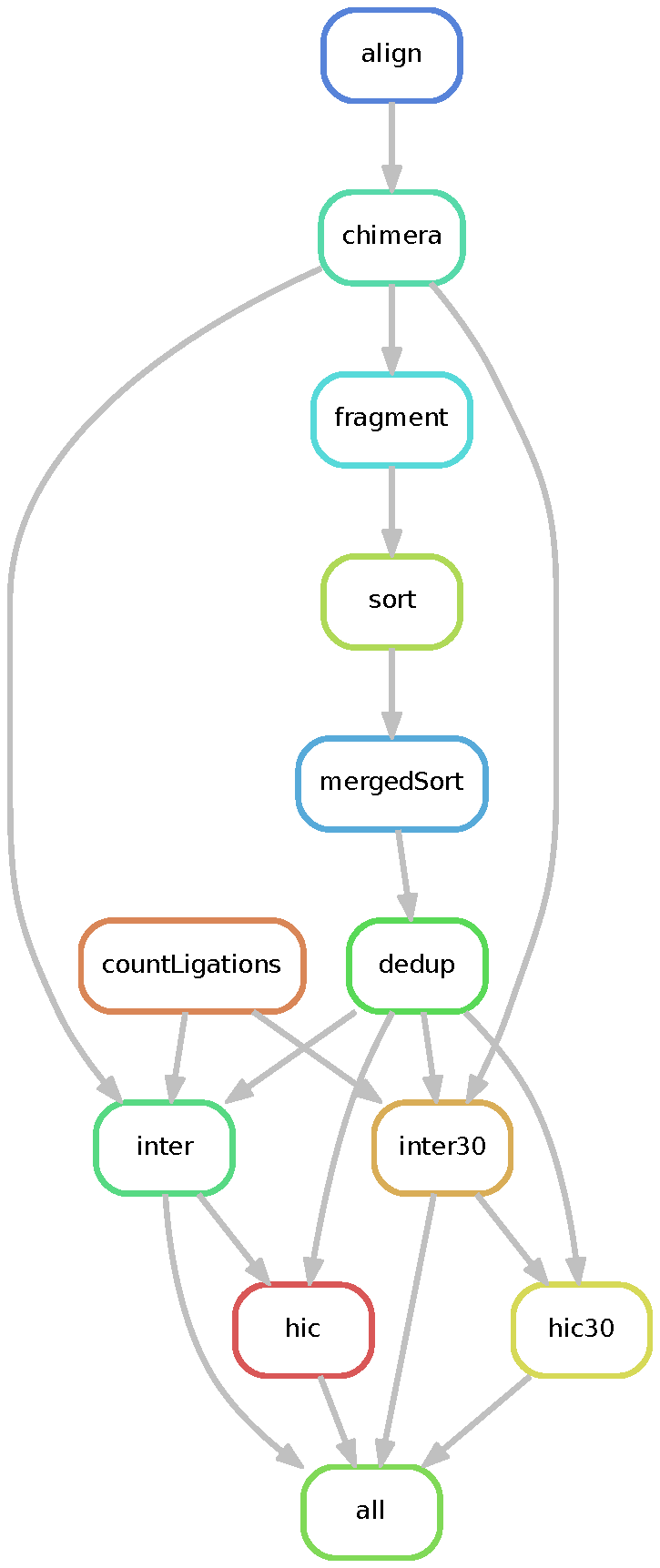
The dietJuicerCore pipeline runs in two stages: 1) splitFASTQ and 2) alignFASTQ. Each stage runs as a separate snakemake workflow, first splitting reads according to the splitsize parameter in the config/config.yaml file, and then following the traditional juicer pipeline steps in alignFASTQ. The pipeline results in stats files, and a merged_nodups file which can be used to create a .hic file.
See the diagrams below for a DAG representation of the workflows:
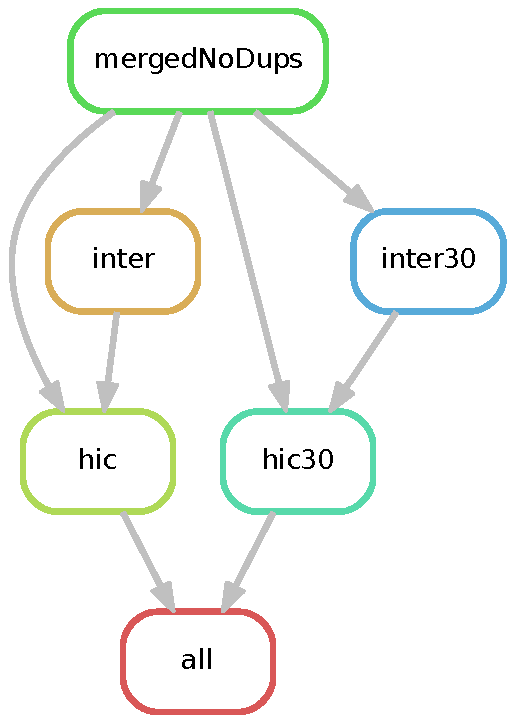
The dietJuicerMerge pipeline implements the buildHIC workflow. It takes output from multiple dietJuicerCore runs and creates a "mega" Hi-C map.
See the diagram below for a DAG representation of the workflow:
dietJuicer uses snakemake version 5.10.0. See requirements.txt file for a list of python dependencies. Using the shell scripts to launch dietJuicer in a cluster setting will automatically run the pipeline in a python virtual environment with the required dependencies.
After dietJuicer successfully runs, you can check the quality of your samples by looking at each inter_30.txt file. Instead of looking at each individual file, you can run the the /scripts/juicerSummary.R script to create a single file that contains all of the information from each inter_30.txt file. From the cloned directory, carry out the following steps:
-
Load R module:
module add r
-
Launch the script:
Rscript ./scripts/juicerSummary.R
The script will generate a file juicerSummary.txt in the directory from which it was launched that contains 29 rows and as many columns as inter_30.txt files found.
If you are interested in seeing how long certain steps took on a successful dietJuicer run, you can run the scripts/benchmarking.py script by carrying out the following steps:
-
Load python module:
module add python
-
Launch the script:
python ./scripts/benchmarking.py
This script will generate a file benchmarking.tsv in the directory from which it was launched that provides valuable information about the computational resources/runtime required by different steps in the pipeline.
Occasionally the pipeline may fail or terminate prematurely. Usually this is due either to a misspecification of job resources (such as insufficient memory, or too short of a runtime) or to changes in the UNC longleaf cluster. You can check the progress of your dietJuicer run by viewing the outfile (e.g. dietJuicerCore-<JOBID>.out). Errored jobs will be reported in this log file, as it collects the output from all steps in the pipeline. For more specific information on the error, navigate to output/logs_slurm and output/{group}/logs.
One of the good things about dietJuicer is that since it is written using snakemake, it will automatically pick up the pipeline right where it left off. So there is no "wasted" time having to restart a stalled pipeline from scratch.
If the error is caused by a misspecification of resources, the user can adjust the resource for the jobs in question by editing the config/cluster.yaml file. While the default values may serve most situations, running particularly large datasets may require some adjustment. Benchmarking successful runs using the scripts/benchmarking.py script can provide an idea of the computational resources required by different steps in the pipeline, given a particular filesize.
If the error is due to job scheduler errors (e.g. slurm on UNC's longleaf cluster), recognizable by sacct or other slurm references in the dietJuicer-out file, then no changes need to be made to the pipeline. Carry out the following steps to get the pipeline back to processing from where it left off:
-
Unlock the directory:
For a failed
dietJuicerCoreworkflow use the unlock script with either "alignFASTQ", "dietJuicerCore" or "Core" as the first argument:./unlock.sh alignFASTQ
If the workflow failed during splitting (an unlikely scenario) - then create a new folder and start the run again from scratch.
For a failed
dietJuicerMergeworkflow use the unlock script with either "buildHIC", "dietJuicerMerge" or "Core" as the first argument:./unlock.sh buildHIC
snakemakeautomatically locks directories to prevent accidental overwrites of files and to prevent multiple incarnations of the program from writing to the same files. Theunlock.shscript unlocks the directory using the correct version of snakemake by activating the python virtual environment. -
Relaunch the pipeline:
For dietJuicerCore:
sbatch dietJuicerCore.sh
For dietJuicerMerge:
sbatch dietJuicerMerge.sh
You should see jobs begin to relaunch within ~1 min after invoking this command.
One caveat is that the pipeline will not automatically remove some unneeded files if it was relaunched. Therefore, users may need to manually delete some of the excess intermediate files (those not listed as outputs above).
If an error persists after taking these steps, it is possible that there is an error in the pipeline. Please document and report these occurances under the issues github page.