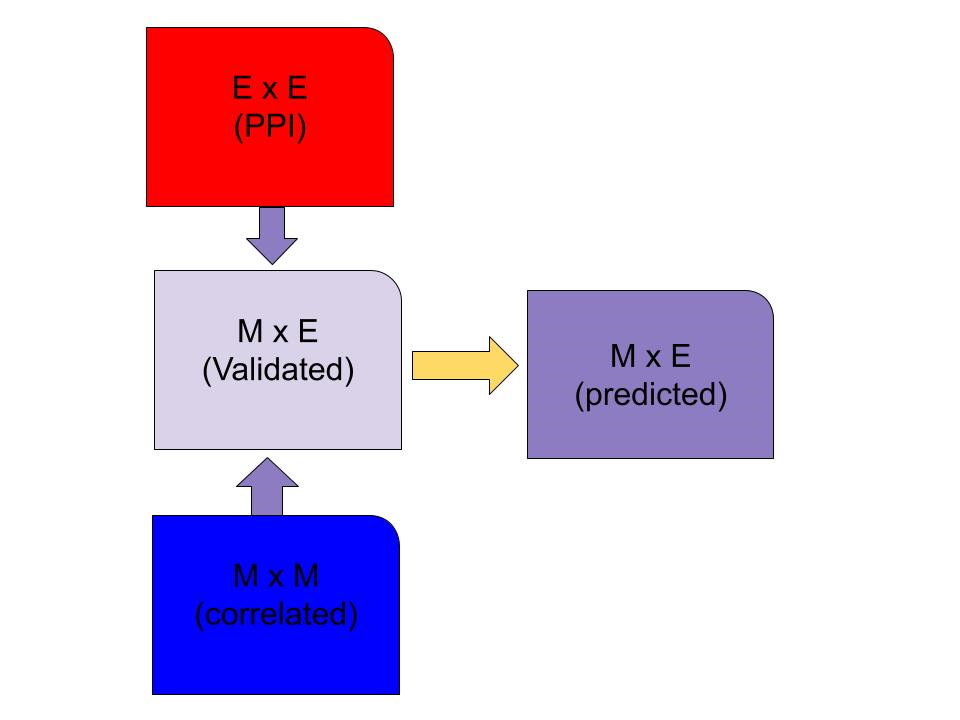
MEAPis is a new algorithm MeaP (Metabolite enzyme association Predictor), which predicts the potential associations between
metabolites and enzymes in metabolomics data. MeaP uses the enzyme-metabolite interaction data from Kyoto Encyclopedia of Genes and Genomes (KEGG) reaction database and the protein-protein interaction data from STRING database.
It used panda function implemented in PandaR R package. https://www.bioconductor.org/packages/release/bioc/html/pandaR.html
You need three matrices to run Meap:
1- Metabolites-Metabolites correlation (M X M)
2- Metabolite enzyme interaction from KEGG (M X E)
3- Protien Protein interaction (E X E)
Here is an example of how to use Meap:
https://nbviewer.jupyter.org/github/FADHLyemen/MEAP/blob/master/MeaP.ipynb