problem and objectives
Study the existance of the micriobiome community inside the placenta. We examined the placental microbiome in a multi-ethnic maternal obesity cohortmethods
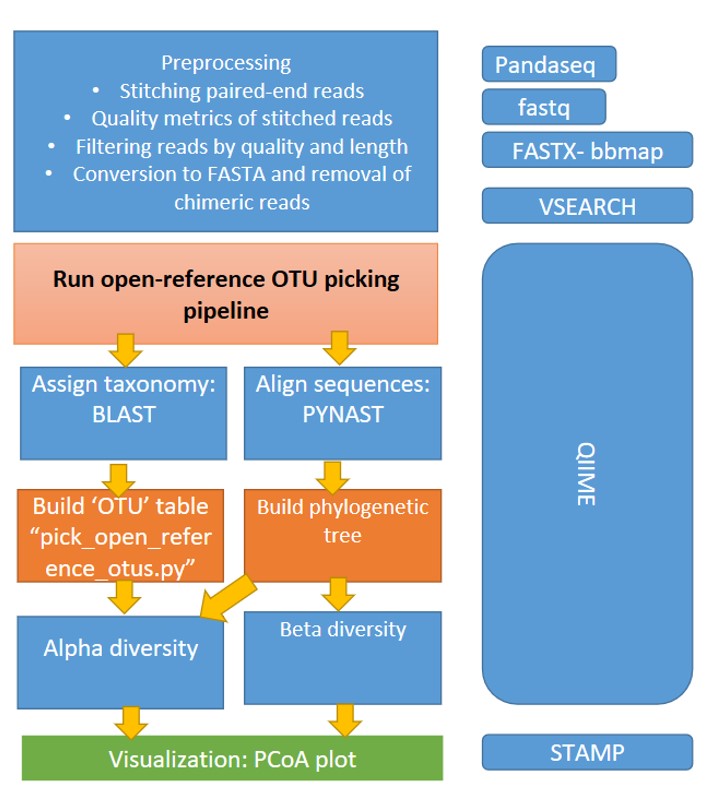
Detailed bioinformatics pipeline used to analyze 16S reads in this study. The 16S rRNA gene reads were analyzed using the pipeline shown in Supplementary Figure 1. Reads were stitched using Pandaseq using 150 and 350 as minimum and maximum length of the assembled reads respectively. Operational taxonomic units (OTUs) were created by clustering the reads at 97 % identity using UCLUST. Representative sequences from each OTU were aligned using PyNAST , and a phylogenetic tree was inferred using FastTree v. 2.1.3 after applying the standard Lane mask for 16S rRNA gene sequences. Pairwise UniFrac distances were computed using QIIME , and permutational tests of distance and principal coordinates analyses were performed with using the MicrobiomeAnalyst, a web-based tool for comprehensive exploratory analysis of microbiome [ref]. Taxonomic assignments were generated by the UCLUST consensus method of QIIME 1.9, using the GreenGenes 16S rRNA gene database v. 13_8results
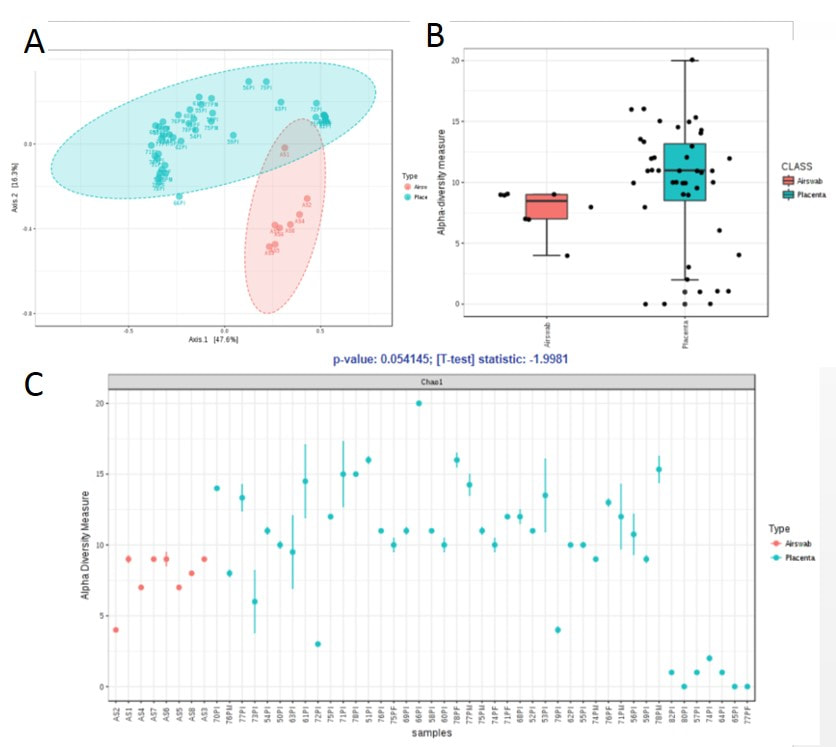
In a PCA plot, we observed that airswabs and placental microbiome were indeed distinct and separable into two clusters.Figure 6: (A) PCA plot comparing obese and non-obese placenta samples. (B-C) Box-plot showing microbial diversity between obese and non-obese placenta samples (p=0.15837, ANOVA F-value=1.9154)
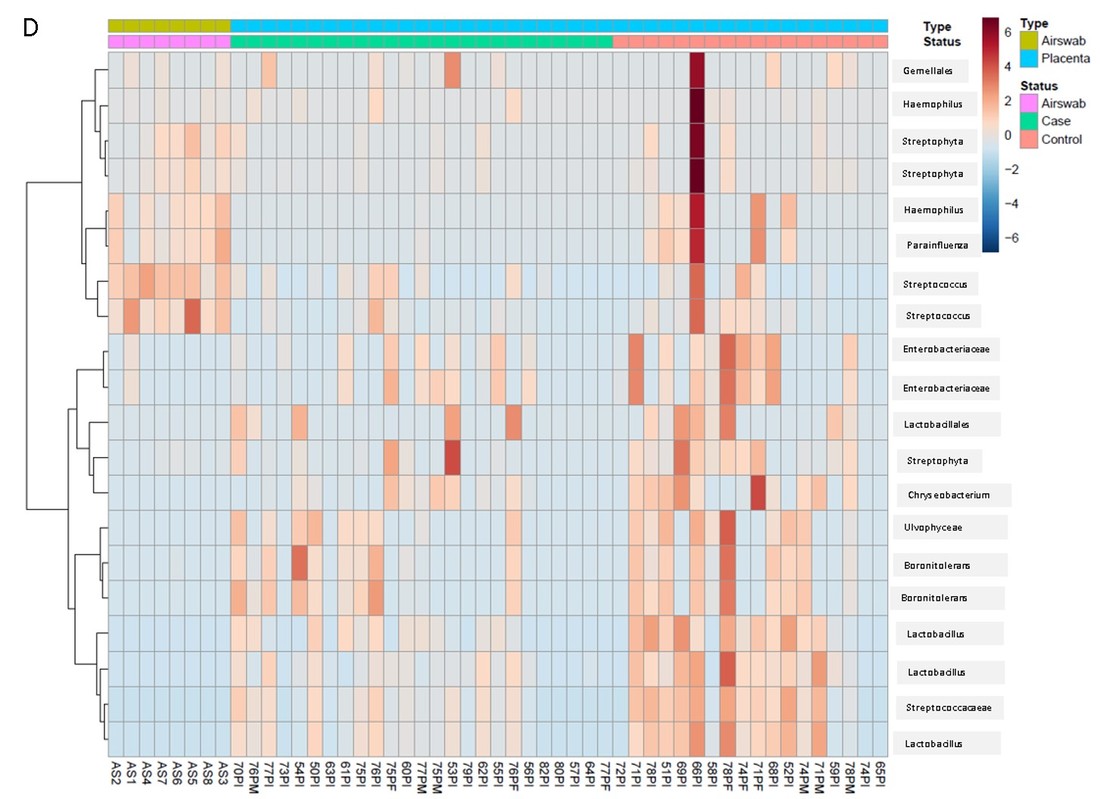
There is a clear difference in bacteria detected as shown in the heatmap.