Download food web datasets from online databases, process, compute structural metrics and evaluate robustness.
DISCLAIMER: This is development version. The code is continuously being subjected to improvements. Any suggestions are welcomed!
In the blog Geekcologist I provided an example of the use of some of these functions: https://geekcologist.wordpress.com/2020/02/06/function-to-download-biotic-interaction-datasets/
library(devtools)
install_github("FMestre1/fw_package")
library(FWebs)
mg1 <- create.fw.list(db="mg", ref=TRUE, spatial = TRUE)
is.adjacency.matrix(mg1)
mg2 <- convert2adjacency(mg1)
is.adjacency.matrix(mg2)
is.sq.matrix(mg1)
mg3 <- rec2square(mg2)
is.sq.matrix(mg3)
metrics <- fw.metrics(mg1)
metrics
dd.fw(mg1, log=TRUE, cumulative=TRUE)
#Convert to list a list of ghraph objects
graph_list1 <- convert.to.graph.list(mg1)
#Create a vector with the values for the Intentionality Index (I)
i_index <- seq(from = 0, to = 1, by =0.01)
i_index <- head(i_index,-1)
#Extract one food web as example
fw1 <- graph_list1[[40]]
#Compute the probability to remove each species
prob_exp <- exponent.removal(fw1, i_index)
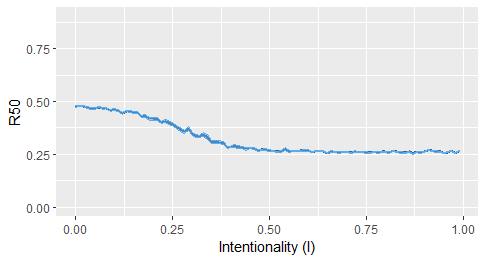
#Simulate the extraction of species to evaluate how many primary extinctions are required to have 50% of the total species extinguished
it1 <- iterate(fw_to_attack=fw1, prob_exp, alpha1=50, iter=10, i_index, plot = TRUE)