SCAGE is a self-conformation-aware pre-training framework for molecular property prediction reveals the quantitate structure-activity relationship like human experts
- Geometry-enhanced molecular representation learning for property prediction
- Accurate prediction of molecular properties and drug targets using a self-supervised image representation learning framework
You can directly use the following command lines to run the code in bash script.
CUDA_VISIBLE_DEVICES=1 nohup python pretrain.py > ./pretrain_log/pretrain1.log 2>&1 &export LD_LIBRARY_PATH="/home2/s439850/anaconda3/envs/SAGE/lib:$LD_LIBRARY_PATH"export LD_LIBRARY_PATH="/home2/s439850/anaconda3/envs/SAGEHH/lib:$LD_LIBRARY_PATH"CUDA_VISIBLE_DEVICES=3 python pretrain.pypython -m torch.distributed.launch --nproc_per_node=4 pretrain_dis.pyunset http_proxy
unset https_proxynnictl create --config ./config.yaml -p 3325close nni and then view it again
nnictl stop --port 3329
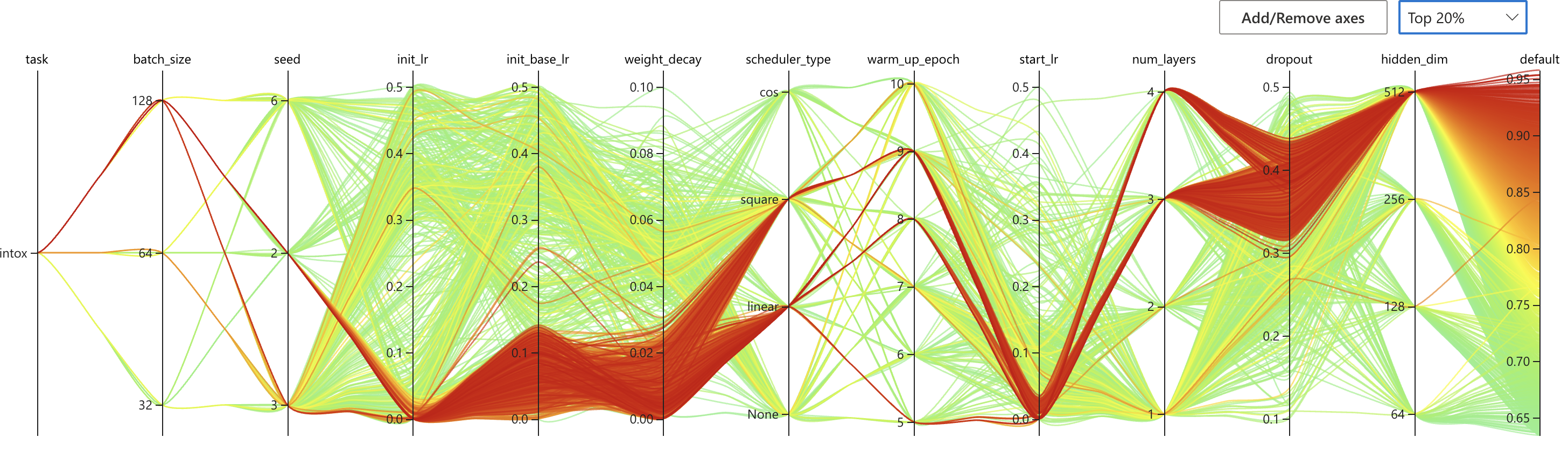
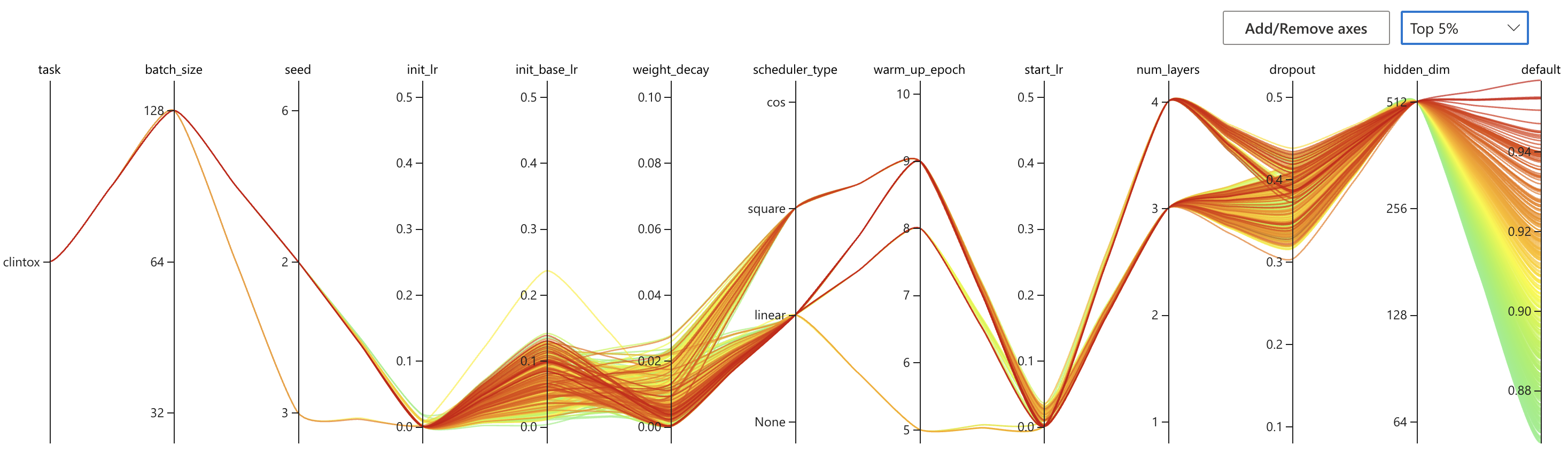
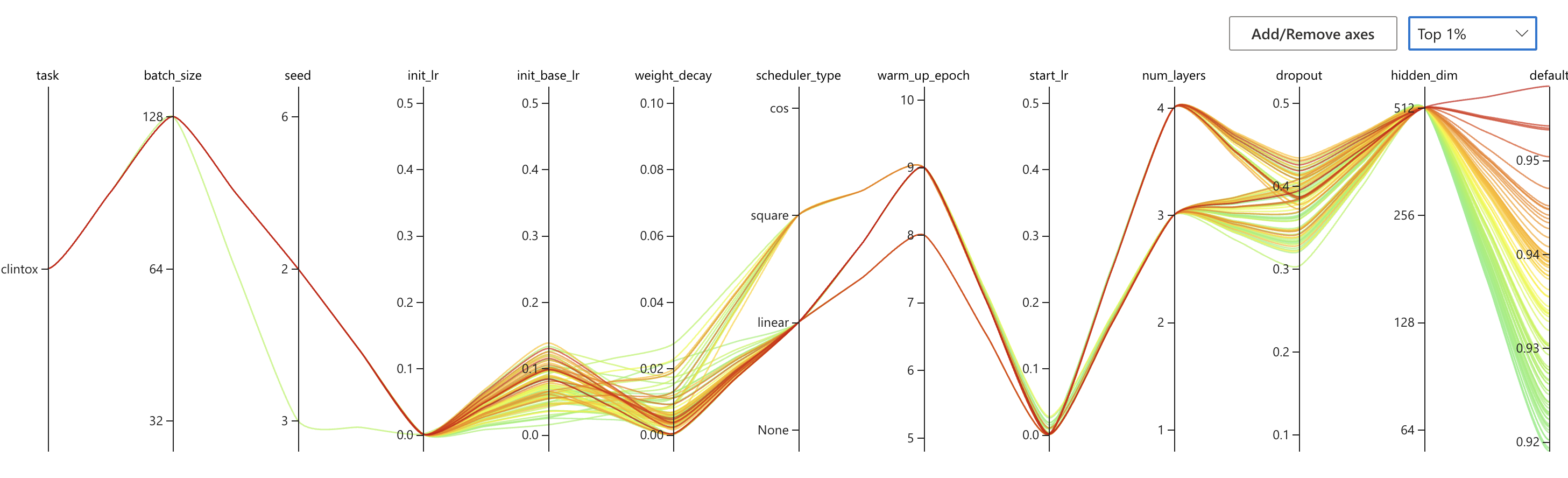
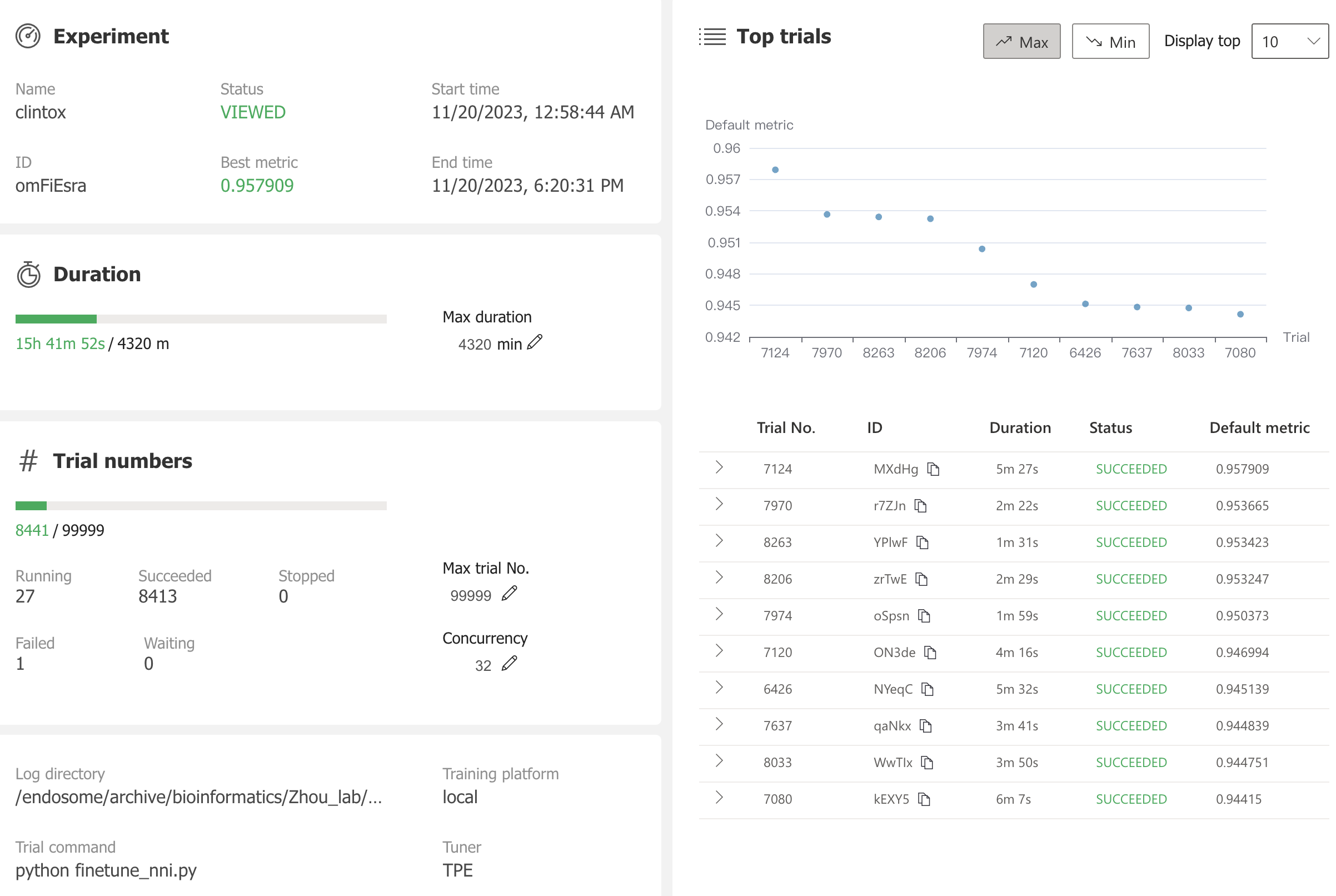
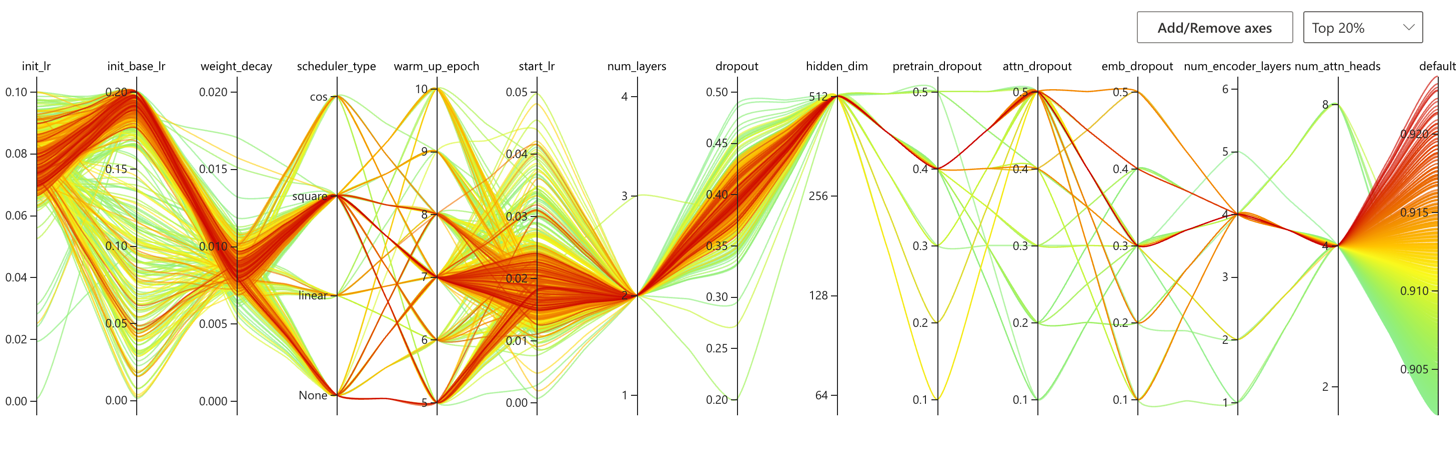
nnictl stop -annictl view omFiEsra --port 3328 -e /archive/bioinformatics/Zhou_lab/shared/jjin/SCAGE/logSome conclusions:
- Choose dist bar as [1,2,3]
- Choose dropout to 0.3/0.4
- Choose number encoder 4
- Choose number layer 4