Python Tool for Synthetic MRI
Explore the docs »
Report Bug
·
Request Feature
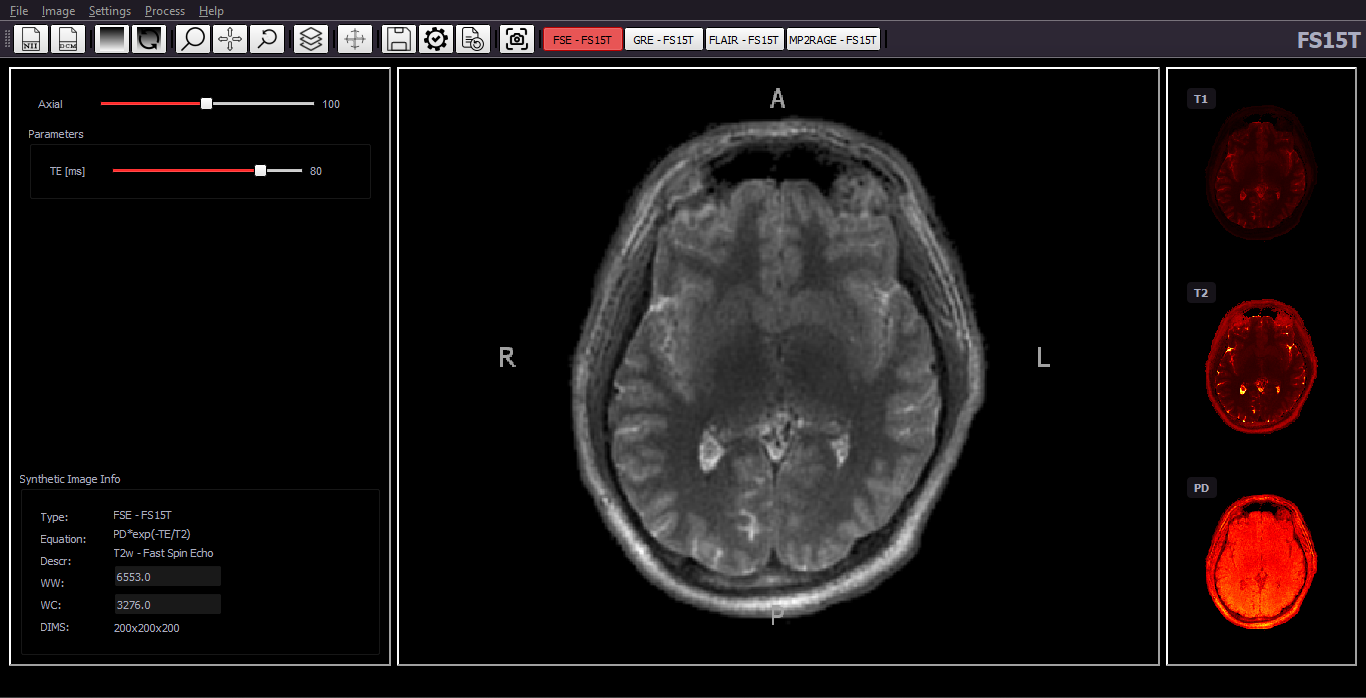 PysynthMRI is a python open-source tool which provides a user-friendly interface to synthesize contrast weighted images. It allows the user to load quantitative maps and adjust virtual scanner parameters to obtain, in real time, the best images.
PysynthMRI is a python open-source tool which provides a user-friendly interface to synthesize contrast weighted images. It allows the user to load quantitative maps and adjust virtual scanner parameters to obtain, in real time, the best images.
If you use this software, please indicate the following reference:
Peretti, L.; Donatelli, G.; Cencini, M.; Cecchi, P.; Buonincontri, G.; Cosottini, M.; Tosetti, M.; Costagli, M. Generating Synthetic Radiological Images with PySynthMRI: An Open-Source Cross-Platform Tool. Tomography 2023, 9, 1723-1733. https://doi.org/10.3390/tomography9050137
- Download Release from: https://github.com/FiRMLAB-Pisa/pySynthMRI/releases/download/v1.0.0/pySynthMRI-v1.0.0-windows-executable.zip
- unzip pySynthMRI-v1.0.0.zip
- run PySynthMRI.exe
These instructions will give you a copy of the project up and running on your local machine,
using Linux or MacOS
Make sure you have Python version >= 3.6.
-
Clone the repository:
git clone https://github.com/FiRMLAB-Pisa/pySynthMRI.git
-
Create a Python Virtual Environment
-
Install required libraries:
pip install -r requirements.txt
In order to correctly execute pySynthMRI, config.json file need to be created.
A default configuration file (config-sample.json) is provided for reference.
We suggest to copy sample file:
cp config-sample.json config.jsonBelow part of the configuration file is reported for help.
{
"synthetic_images": {
"FS15T": { # Name of preset (multiple presets can exists)
"FSE": { # Name of signal model
"title": "T2w - Fast Spin Echo", # Title of signal model
"equation": "PD*exp(-TE/T2)", # Model equation
"parameters": { # List of parameters json object
"TE": { # Name of parameter
"label": "TE [ms]", # Shown label
"value": 80, # Default value
"min": 1, # Min value
"max": 100, # Max value
"step": 1 # Slider/Mouse parameter step
}
},
"series_number": 9001 # [optional] Add to SeriesNumber DICOM tag (0020,0011)
},
[...]
"quantitative_maps": { # Quantitative maps that can be loaded
"T1": { # Name of map
"file_name": "qmap_t1" # substring filename (used if autoload qmaps)
},
"T2": {
"file_name": "qmap_t2"
},
"PD": {
"file_name": "qmap_pd"
}
},
[...]
}You can launch PySynthMRI running launcher.py in your Python IDE or via operating system command-line or terminal:
python pySynthMRI.pyCustom signal models can be added at runtime or using the configuration file.
To facilitate the user, PySynthMRI contains a set of default contrast images in its configuration file:
| Title | Equation | Description |
|---|---|---|
| FSE | PD * ((1 - exp(-TR/T1))*exp(-TE/T2)) | T2w - Fast Spin Echo |
| GRE | PD * ((1 - exp(-TR/T1))*exp(-TE/T2) | T1w - Gradient Echo |
| FLAIR | abs(PD) * exp(-TSAT/T1)*exp(-TE/T2)*(1-2*exp(-TI/T1)) | T2w - Fluid Attenuated Inversion Recover |
| MP2RAGE | 1-2*exp(-TI/T1) | T1w - Magnetization-Prepared 2 RApid Gradient Echoes |
| DIR | PD * (1 - 2*exp(-TI_2/T1) + 2*exp(-(TI_1+TI_2)/T1) - exp(-TR/T1)) * (exp(-TE/T2)) | 3D Double Inversion Revovery |
| TBE | PD * (1-2*exp(-TI/T1)) * (1-exp(-TR/T1)) * exp(-TE/T2) | Tissue Border Enhancement by inversion recovery |
In order to test the software with in-vivo data, we provide downloadable example files at the following link: test_data.zip. The zip file contains 3 quantitative, anonymized maps.
If you use this software, please indicate the following reference:
Peretti, L.; Donatelli, G.; Cencini, M.; Cecchi, P.; Buonincontri, G.; Cosottini, M.; Tosetti, M.; Costagli, M. Generating Synthetic Radiological Images with PySynthMRI: An Open-Source Cross-Platform Tool. Tomography 2023, 9, 1723-1733. https://doi.org/10.3390/tomography9050137
or use the bibetex file cite-pysynthmri.bib provided.
PySynthMRI is distributed under GPL-v3.0 License. See LICENSE.txt for more information.




