A Snakemake workflow for recovery and characterization of plastid genomes from metagenomic datasets. plastiC leverages existing metagenomic tools to identify and characterize plastid genomes starting from metagenomic assemblies.
If you use this tool, please cite:
Cameron, E.S., Blaxter, M.L., Finn, R.D. plastiC: A pipeline for recovery and characterization of plastid genomes from metagenomic datasets [version 1; peer review: 1 approved, 1 approved with reservations]. Wellcome Open Res 2023, 8:475 (https://doi.org/10.12688/wellcomeopenres.19589.1)
plastiC requires the following input files:
- Metagenomic assembly (.fasta) - generated from preferred metagenomic assembly tool (e.g., SPAdes). Assemblies are used to screen for potential plastid sequences. Each sample being analyzed should have it's own folder containing the final assembly file with a shared name. Example of file structure for metagenomic assembly input:
example
├── sample_1
│ └── scaffolds.fasta
├── sample_2
│ └── scaffolds.fasta
├── sample_3
│ └── scaffolds.fasta
├── sample_4
│ └── scaffolds.fasta
└── sample_5
└── scaffolds.fasta
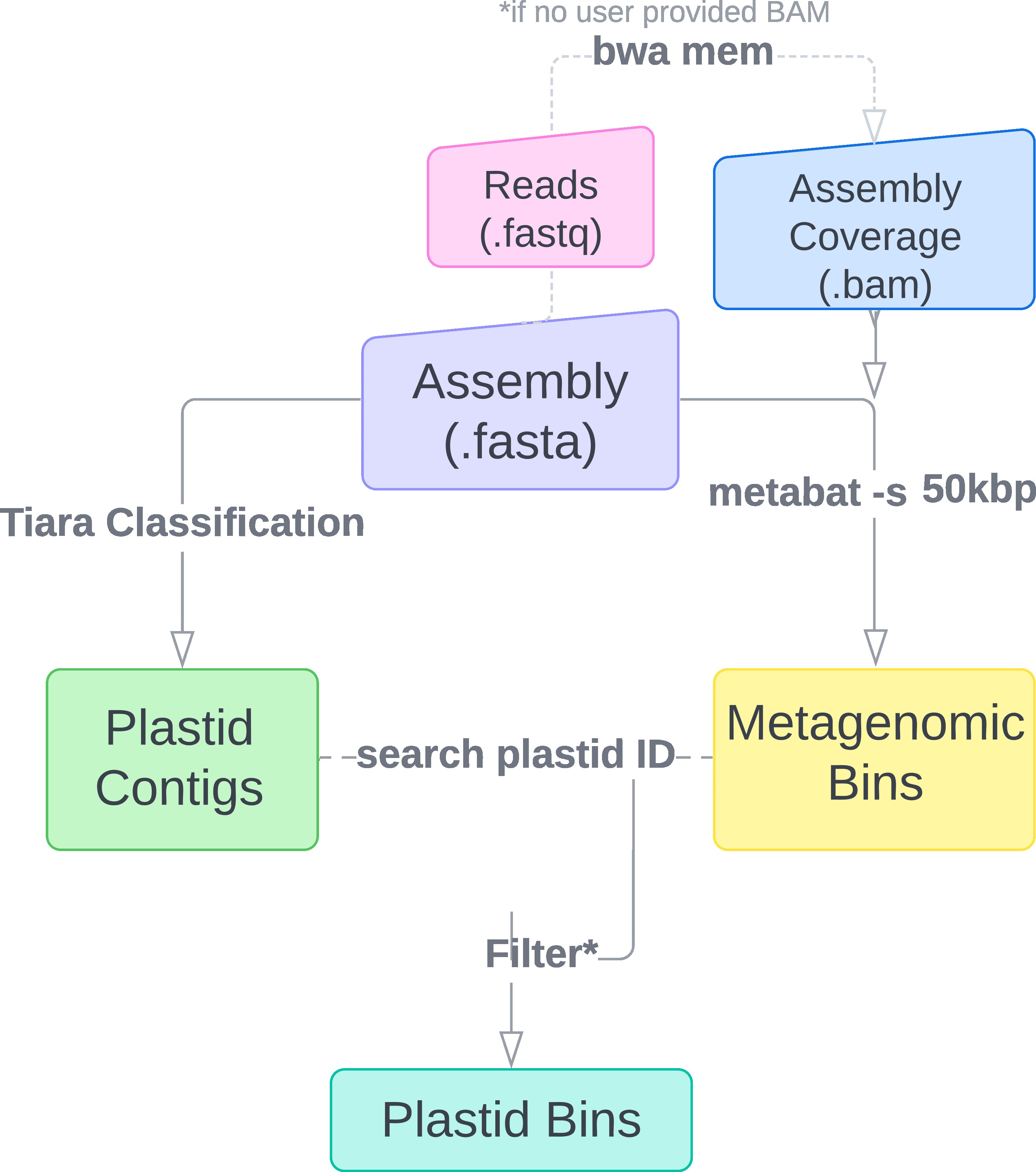
- Metagenomic reads (.fastq) or assembly coverage (.bam) - if available, users can provide an assembly coverage file (.bam) generated from mapping. Alternatively, this coverage file can be generated in the pipeline by providing the metagenomic reads (.fastq). Coverage information is required for generating potential plastid bins.
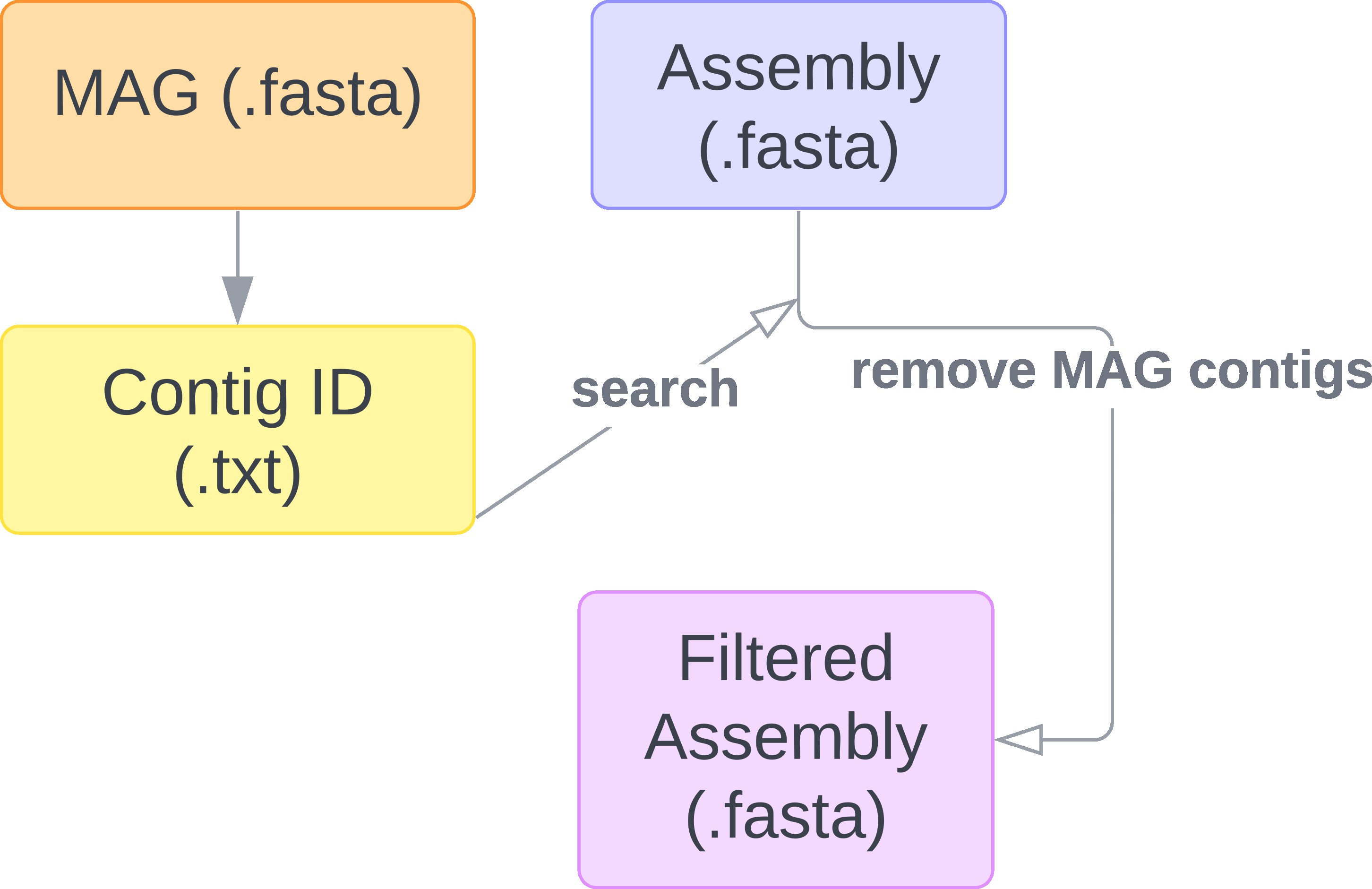
- MAGs (.fasta) - if metagenomic analyses have been conducted on the assembly and high-quality metagenomic assembled genomes (MAGs) for other microbial taxa is available, these can be provided to remove sequences associated with high-quality and high-completeness microbial genomes.
If provided, plastiC initiates by searching metagenomic assemblies for sequence identifiers corresponding to user provided MAG fasta files in the metagenomic assembly. This results in the generation of a filtered assembly fasta file which excludes all sequences which have been previously identified in high-quality MAGs. Filtered assemblies would then be used in the pipeline.
If no MAGs are provided, plastiC will execute on the provided metagenomic assembly fasta file.
Generation of plastid bins requires assembly coverage information. Users may opt to either provide an assembly coverage file (.bam) and skip the mapping step, or to provide paths to read files for plastiC to generate coverage information. Using the coverage information, metagenomic bins are generated from the assembly file using metabat with a lower bin threshold size of 50kbp to account for the small plastid genome size. Concurrently, Tiara is used to identify potential plastid contigs in the assembly.
Sequence identifiers of flagged plastid contigs are used to to search for metagenomic bins containing plastid signals. Plastid bins are selected based on the percentage of the bin coming from plastid sequence. By default this threshold is set to 90% but users can adjust this threshold to their requirements in the config.yaml.
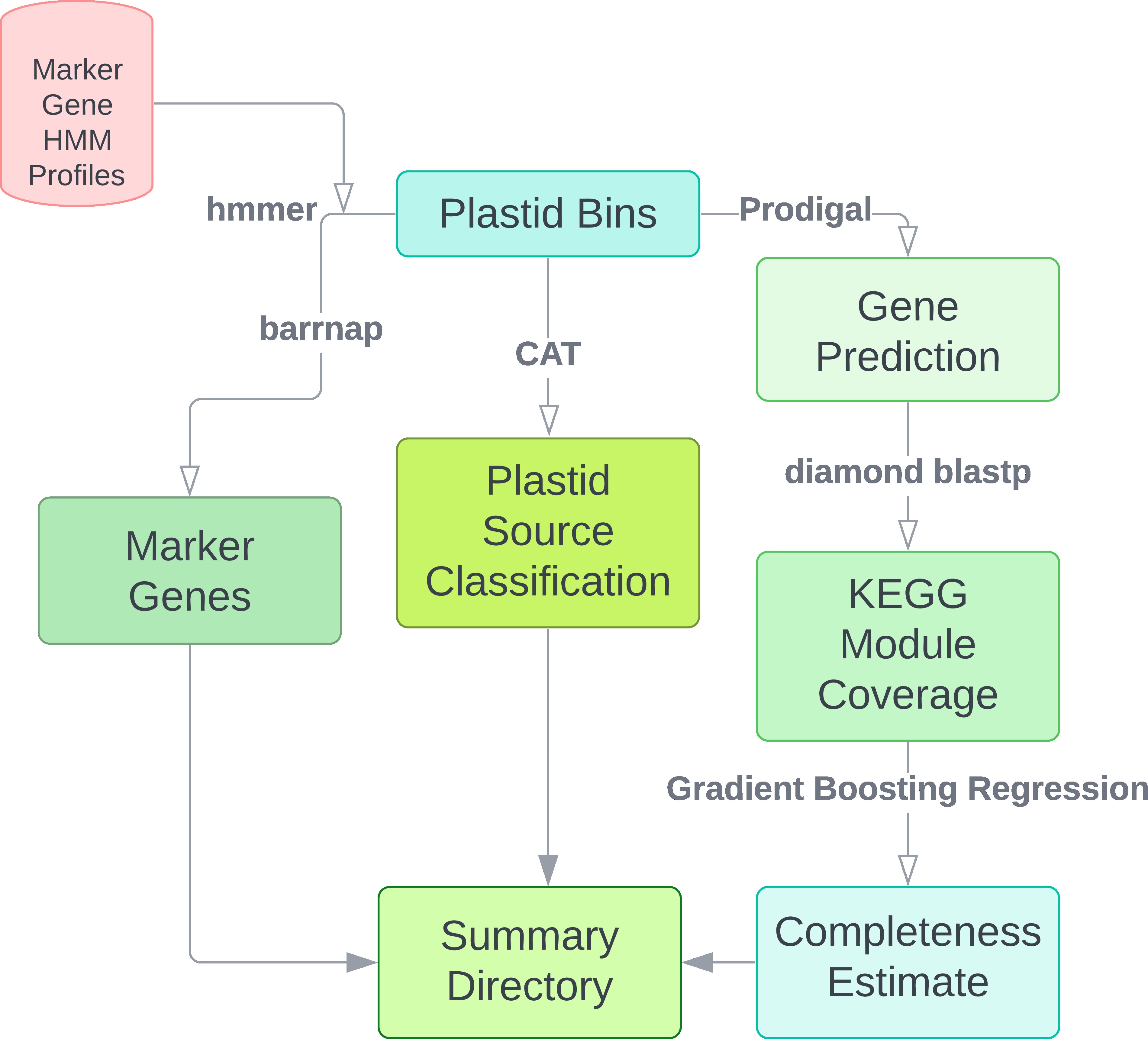
Following generation of plastid bins, plastiC will perform additional analyses to further summarize details of the recovered plastid genomes including taxonomic placement and a completeness estimate. The final completeness estimate of the recovered plastid genomes is reference independent. Users may opt to further explore the plastid genome quality by mapping to reference genomes arising from the same lineage.
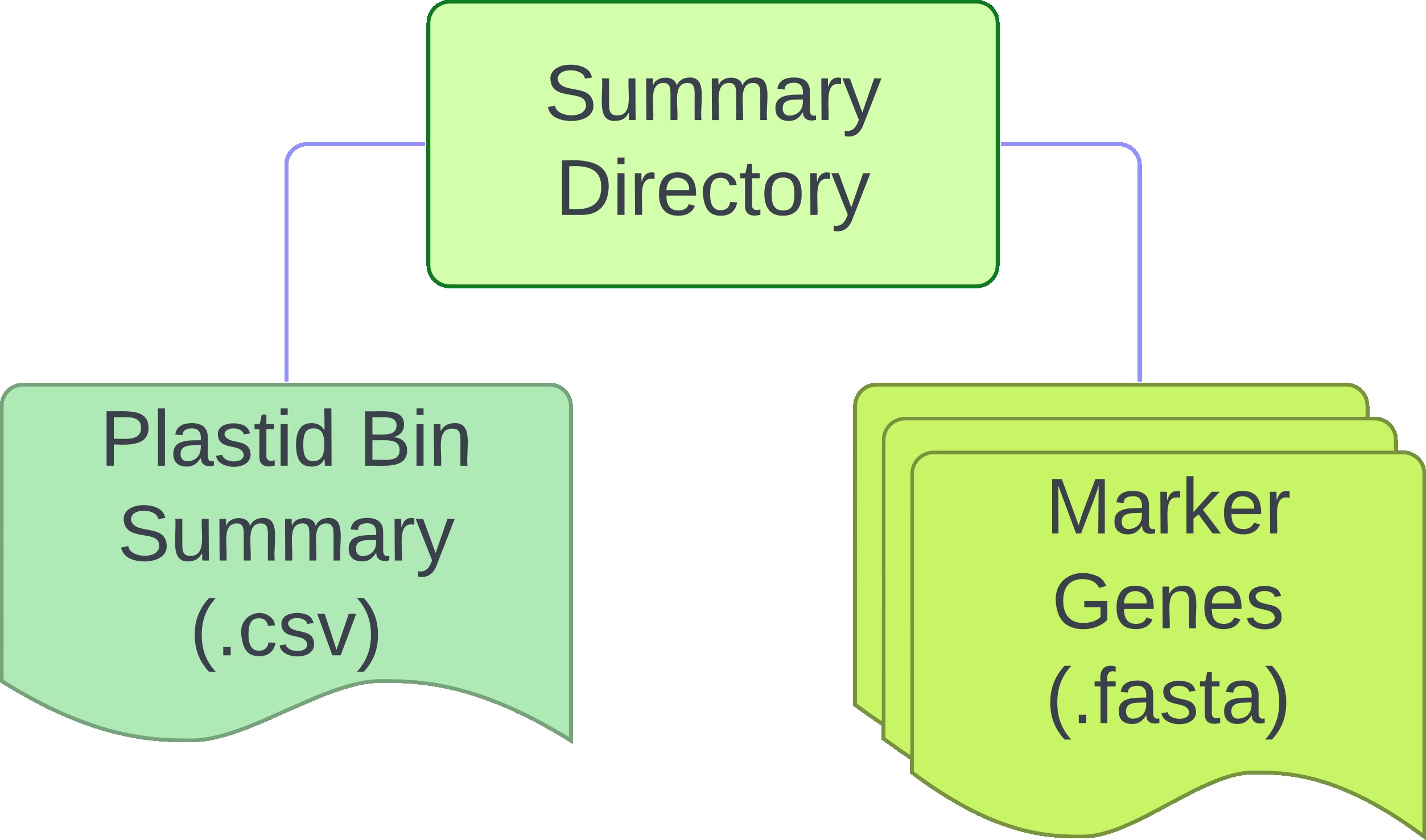
For each sample, three dictories will be created: logs, working and plastids. Final outputs regarding
plastid information can be found in the plastids directory. The working directory contains intermediate files
from analyses required for identification and characterziation of plastid genomes but can be removed if users desire.
plastiC is a Snakemake workflow and to run requires a Snakemake installation (https://snakemake.readthedocs.io/en/stable/getting_started/installation.html). Snakemake can be easily installed using conda as highlighted below. If not already installed, install conda.
Following [conda] installation, Snakemake can be installed into a new environment.
- Create a new environment with Snakemake.
Example:
conda create -n snakemake -c bioconda snakemake
To have access to the workflow, clone the plastiC repository.
2. Clone plastiC repository.
Example:
git clone --recursive https://github.com/escamero/plastiC.git
- Download databases (described below).
The plastiC workflow uses dockerhub to fetch all required tools, so ensure Singularity is also installed.
plastiC requires databases for taxonomic source classification using CAT and for completeness estimation.
Please download the following and provide links to the paths to these databases in the as instructed config.yml.
CAT Databases
Please visit https://github.com/dutilh/CAT for database installation and preparation instructions provided.
Uniref
For completeness estimation, gradient boosting regression models were trained on a diamond database created from the Uniref100 (released November 26, 2018) database with KO annotations as used in CheckM2.
Currently, the workflow can be run using the diamond database created by CheckM2 developers available to download here.
An updated database for completeness estimates will be hosted and released in the future.
Please fill in the config.yaml with file and directory paths as described in the file.
After the above steps have been completed, samples can now be run to identify plastid genomes. The first step is to activate your Snakemake conda environment that was created during setup.
Example:
conda activate snakemake
The value for -j should be adjusted to reflect the number of cores available. The -k flag may be removed if users desire the workflow to stop if an independent job fails. The --use-singularity flag is required for as tools and dependencies for rules are in a docker container.
Example:
snakemake --use-singularity -k -j 2
plastiC can also be executed on a cluster. The specifications (e.g., memory, cores) for cluster execution can be adjusted in the cluster.yml file.
Note: Exact submission command may need to be adjusted depending on your system.
Example (LSF submission):
snakemake --use-singularity -k -j 2 --cluster-config cluster.yml --cluster 'bsub -n {cluster.nCPU} -M {cluster.mem} -o {cluster.output}'