The 15th place solution code for a Kaggle competition, Human Protein Atlas - Single Cell Classification
The task of this competition is a weakly instance segmentation for protein class like below.
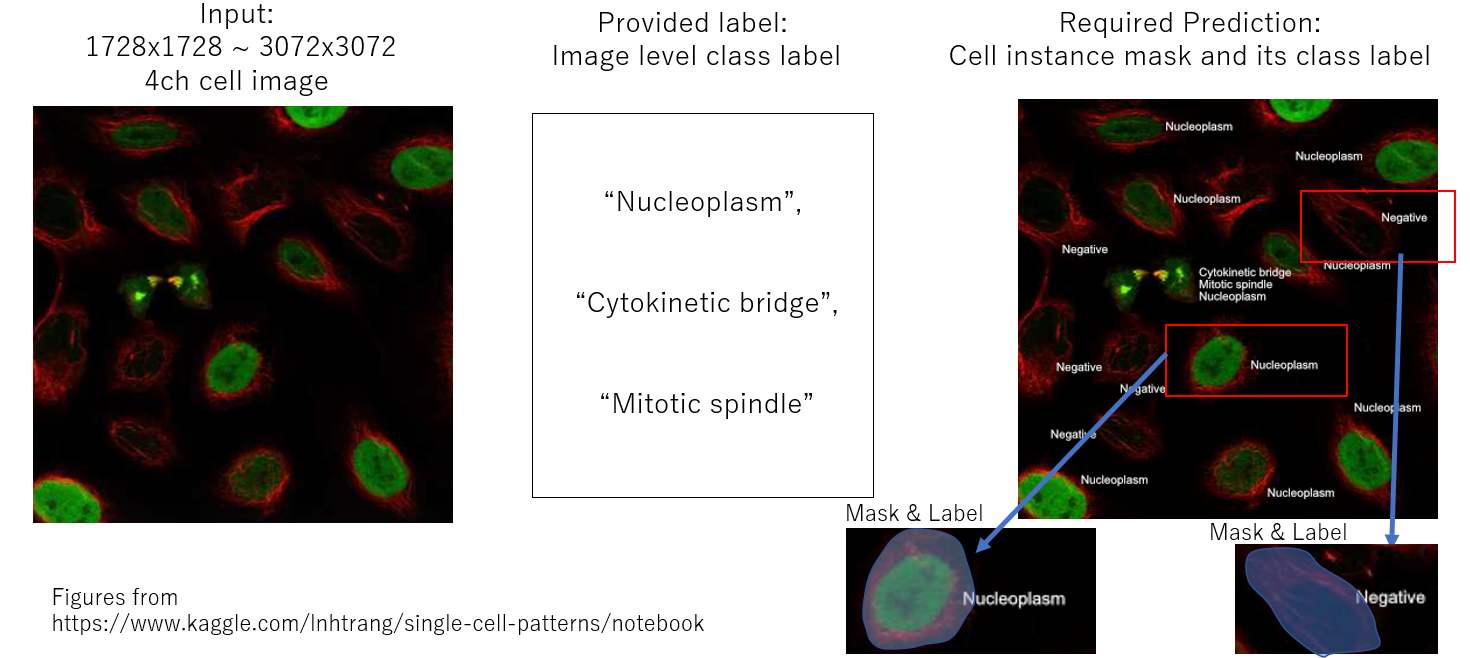
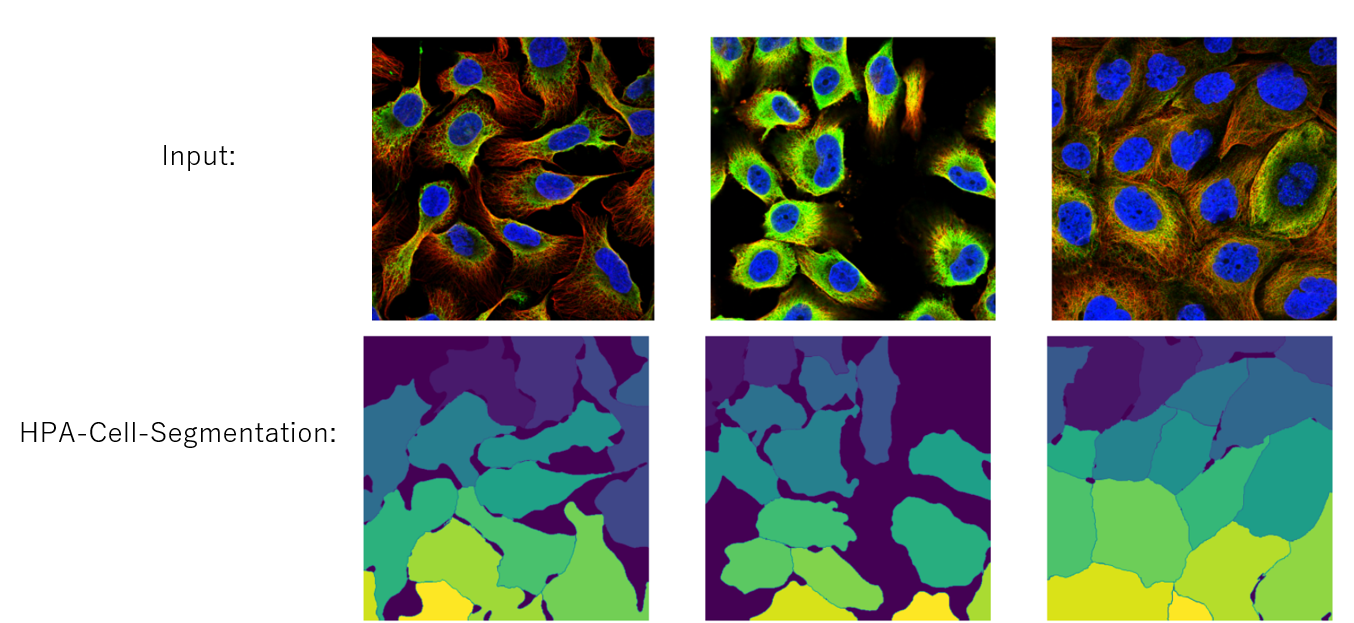
But we have a tool, HPA-Cell-Segmentation, which segments each cell as instance mask. That's why we can only predict class for each mask if we rely on this tool.
 .
.
2 stages approach,
- 1 st stage: CAM x SC-CAM generation
- 2 nd stage: Semantic Segmentation with the pseudo label from CAM
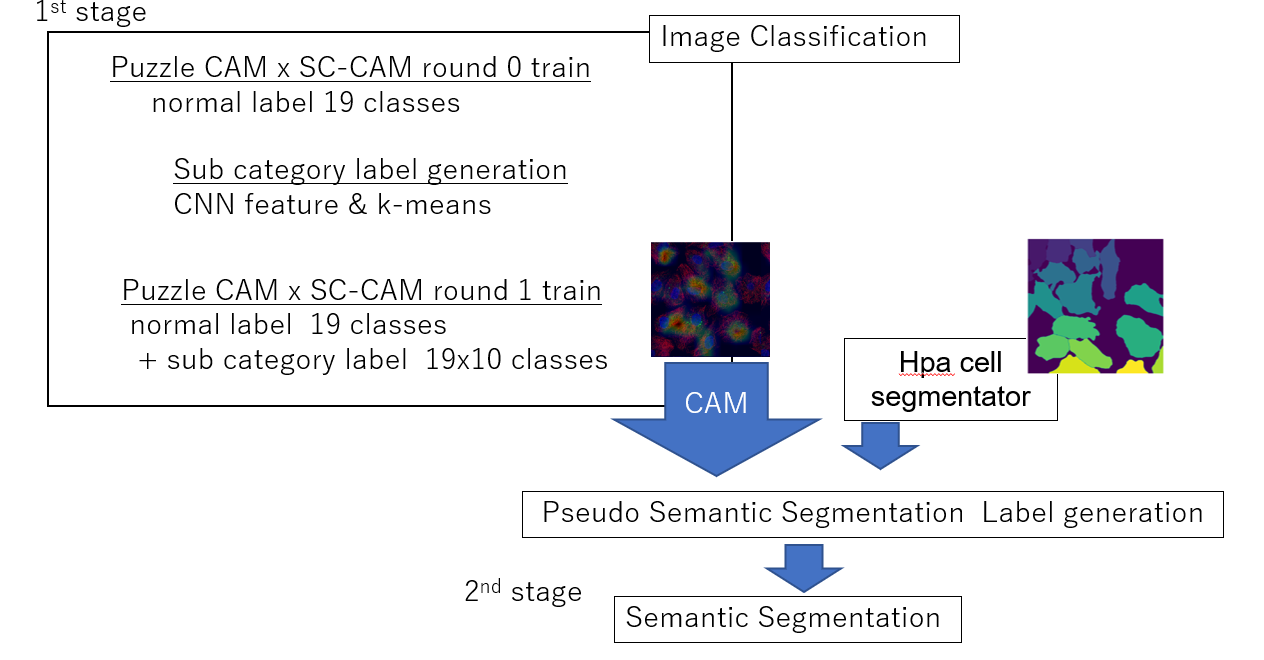
- mAP 0.518, 15th place on this competition.
- This is my kaggle submission notebook.
For more details, please check my slide.
First, download the data, here and if you want to use additional data you need this data and this csv. For hpa segmentation tool, pretrained weights are needed. You can get these weights, here. To run this code, the following configuration is expected.
../input/
│
├ hpa-single-cell-image-classification/
├ hpa-challenge-2021-extra-train-images/
├ publichpa-withcellline_processed.csv
└ hpa-cell-seg-weights
├ nuclei-model.pth
└ cell-model.pth# clone project
git clone https://github.com/Fkaneko/kaggle_2021_hpa_single_cell_classification
# install project
cd kaggle_2021_hpa_single_cell_classification
pip install -r requirements.txt
pip install https://github.com/CellProfiling/HPA-Cell-Segmentation/archive/master.zipLOG_DIR="./"
NUM_WORKERS=16
TIMM=resnet50
EPOCHS=1
DATA_DIR=../input/hpa-single-cell-image-classification
INPUT_SIZE=512
# 1st SC-CAM round 0
EXT_MODE=0
ROUND_NB=0
python run_classification.py \
--gpu 1 \
--max_epochs $EPOCHS \
--val_fold 0 \
--batch_size 40 \
--data_dir ${DATA_DIR} \
--timm_model_name $TIMM \
--benchmark \
--precision 16 \
--optim_name sgd \
--round_nb ${ROUND_NB} \
--aug_mode 1 \
--default_root_dir ${LOG_DIR} \
--num_workers ${NUM_WORKERS} \
--num_inchannels 4 \
--use_ext_data \ # assume using extra data
--ext_data_mode ${EXT_MODE} \
--input_size ${INPUT_SIZE} \
--lr 0.5After this SC-CAM round 0 training, for round 1, using the trained weight of round 0, sub-category is generated and moving on to round 1 like below.
ROUND_NB=1
CKPT_PATH=`ls ${LOG_DIR}/lightning_logs/version_0/checkpoints/epoch*` # need to change
SUB_LABEL_DIR="./save_uni"
mkdir $SUB_LABEL_DIR
python ./extract_feature.py \
--weights $CKPT_PATH \
--save_folder ${SUB_LABEL_DIR} \
--use_ext_data \ # assume using extra data
--ext_data_mode ${EXT_MODE}
python ./create_pseudo_label.py \
--save_folder ${SUB_LABEL_DIR} \
--for_round_nb $ROUND_NB \
--is_scale_feature \
--use_ext_data \ # assume using extra data
--ext_data_mode ${EXT_MODE}
# 1st SC-CAM round 1
python run_classification.py \
--gpu 1 \
--max_epochs $EPOCHS \
--val_fold 0 \
--batch_size 40 \
--data_dir ${DATA_DIR} \
--timm_model_name $TIMM \
--benchmark \
--precision 16 \
--optim_name sgd \
--round_nb ${ROUND_NB} \
--aug_mode 1 \
--sub_label_dir ${SUB_LABEL_DIR} \
--default_root_dir ${LOG_DIR} \
--num_workers ${NUM_WORKERS} \
--num_inchannels 4 \
--use_ext_data \ # assume using extra data
--ext_data_mode ${EXT_MODE} \
--input_size ${INPUT_SIZE} \
--lr 0.005After 1st stage, moving on to pseudo semantic segmentation label generation and segmentation training.
But at this pseudo label generation, it will take ~70h ? I guess.
Because hpa segmentation tool is not fast. After the initial run, masks of hpa segmentation tool
are going to be cached at ../input/hpa_mask/ and the processing time will reduce to ~10h.
# pseudo label generation for 2nd stage
SEGM_DIR=$SUB_LABEL_DIR
python run_cam_infer.py \
--yaml_path ./src/config/generate_pseudo_label.yaml \ # you have to change `ckpt_paths` at this yaml
--sub_path ${SEGM_DIR}/pseudo_label.csv
python create_segm_labels.py \
--save_folder ${SEGM_DIR} \
--sub_csv ${SEGM_DIR}/pseudo_label.csv
# 2nd stage semantic segmentation
SEGM_TRESH=0.225
TIMM="resnet50"
INPUT_SIZE=512
python run_classification.py \
--gpu 1 \
--max_epochs $EPOCHS \
--val_fold 0 \
--batch_size 12 \
--benchmark \
--precision 16 \
--timm_model_name ${TIMM} \
--optim_name sgd \
--segm_label_dir $SEGM_DIR \
--segm_thresh $SEGM_TRESH \
--round_nb 0 \
--num_workers $NUM_WORKERS \
--default_root_dir $LOG_DIR \
--num_inchannels 4 \
--data_dir ${DATA_DIR} \
--use_ext_data \ # assume using extra data
--ext_data_mode ${EXT_MODE} \
--input_size ${INPUT_SIZE} \
--lr 0.25The label to test data is hidden so we could not evaluate a model locally.
We have to submit our model on a kaggle server.
I have used run_cam_infer.py for evaluation at kaggle and I shared it here.
If you want to evaluate your trained model, please change ckpt_paths of src/config/kaggle_submission.yaml.
Please check my kaggle post for details.
This is my kaggle submission and got, mAP 0.518, 15th place on this competition. If you want to reproduce my training results, the easiest way is getting hyperparamters from classification models and semantic segmentation models. After downloading these datasets you can get all hyperparameters for each model like below and these hyperparamters can be used for reproducing the same training configuration.
from src.modeling.pl_model import LitModel, load_trained_pl_model
checkpoint_path = "lightning-weights/rst50_1024_r0_ext1_val1/version_68/checkpoints/epoch-24-step-82924.ckpt"
_, args_hparams = load_trained_pl_model(LitModel, checkpoint_path=checkpoint_path, only_load_yaml=True)Also each hyperparameter is checked with tensorboard. But there are so many parameters, it's hard to check with tensorboard...
# download https://www.kaggle.com/sai11fkaneko/lightning-weights
tensorboard --logdir ./lightning-weights
# now starting tensorboard, and `HPARAMS` tab is availableIf you reproduce my training result, you need download external data, which is not available as kaggle dataset. So you can use download_ext_data.py. But it takes 5 or 6 days.
MIT
From https://www.kaggle.com/c/hpa-single-cell-image-classification/rules,
DATA ACCESS AND USE: COMPETITION AND ACADEMIC/RESEARCH USE ONLY
And the license of my pretrained weights are supposed to be the same as the dataset license.

