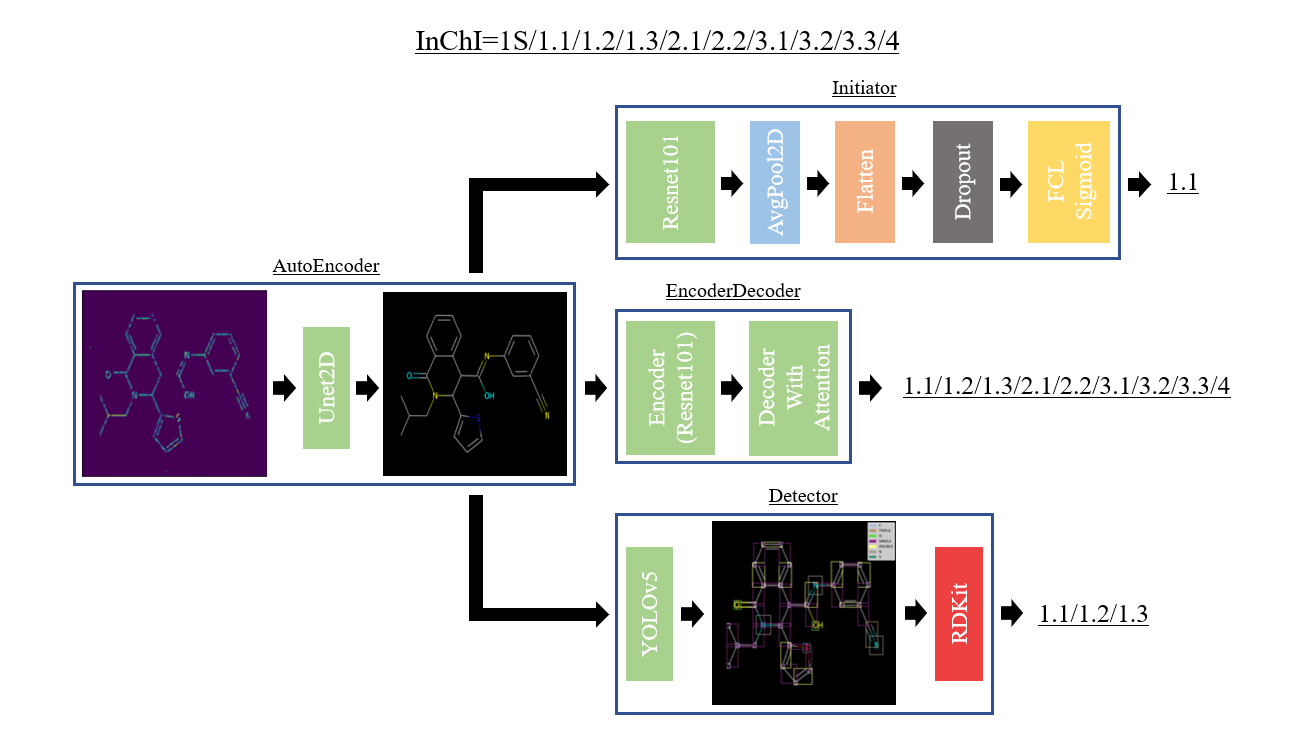
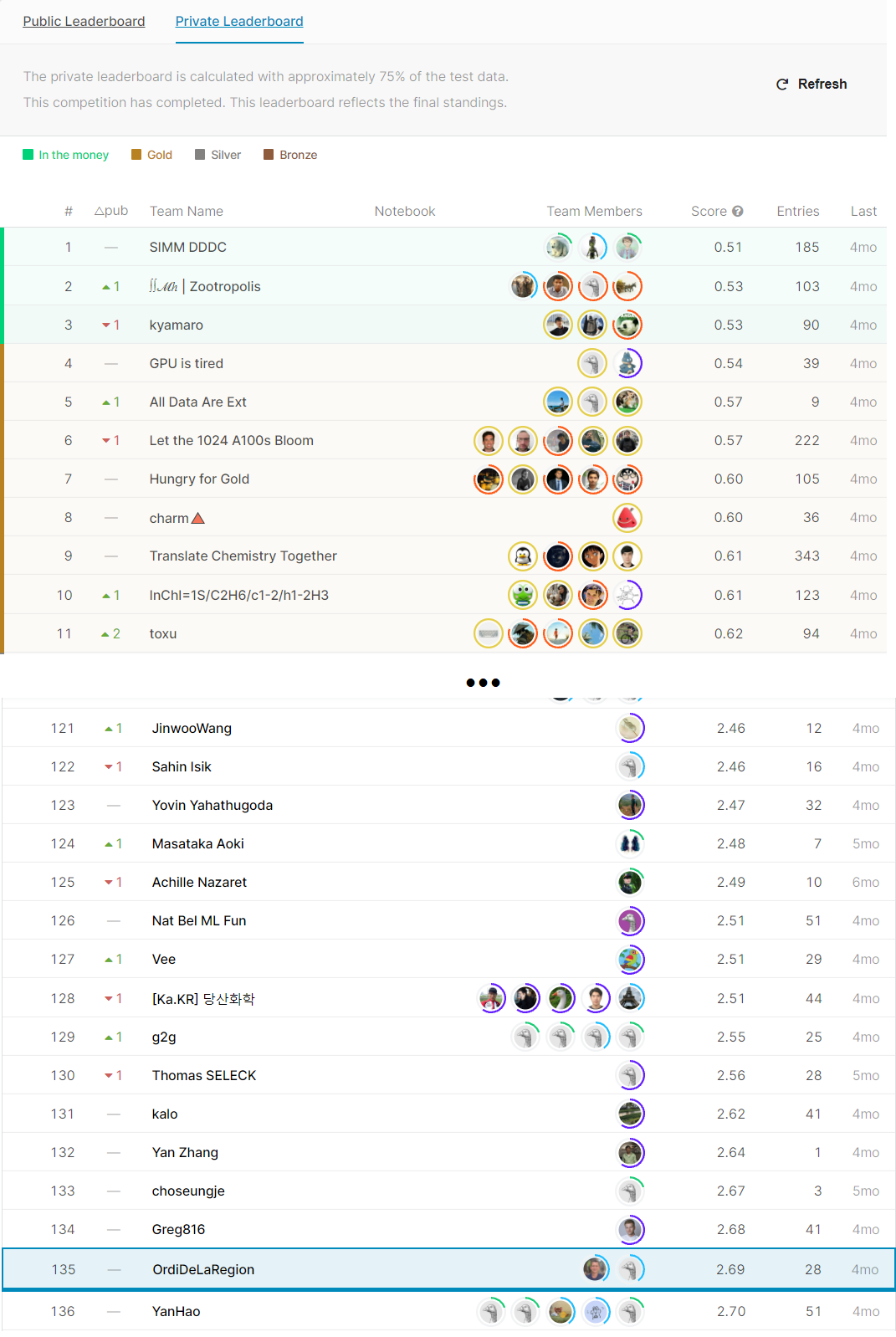
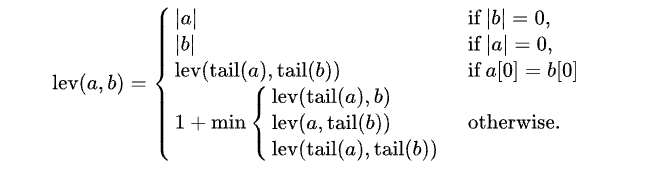This repository presents the solution developed during the BMS-Molecular-Translation competition. This one is composed of four parts:
- AutoEncoder
- Detector
- EncoderDecoder
- Initiator
These four parts were assembled in the final submission to provide an innovative and original solution 👍
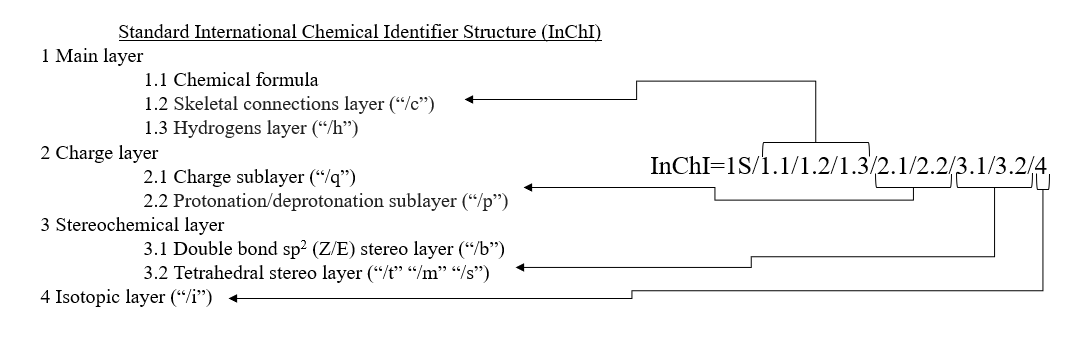
International Chemical Identifier Structure (InChI)
- Automated recognition of optical chemical structures
- Convert images back to the underlying chemical structure (InChI text)
- Help chemists expand access to collective chemical research
"Levenshtein Distance is defined as the minimum number of operations required to make the two inputs equal. Lower the number, the more similar are the two inputs that are being compared." (Devopedia, 2021)
InChI is a non-proprietary, Open Source, chemical identifier intended to be an IUPAC approved and endorsed structure standard representation.
Features of chemical structure in a hierarchical, layered manner. Major InChI layers: Main, Charge, Stereo, Isotopic, FixedH (never included in standard InChI) as well as the Reconnected layer (never included in standard InChI), and their associated sublayers.
This section was built with: Heller, S.R., McNaught, A., Pletnev, I. et al. InChI, the IUPAC International Chemical Identifier. J Cheminform 7, 23 (2015). https://doi.org/10.1186/s13321-015-0068-4
Architecture of the model with the outputs associated to each branch. The final prediction is a combination of the three branches.
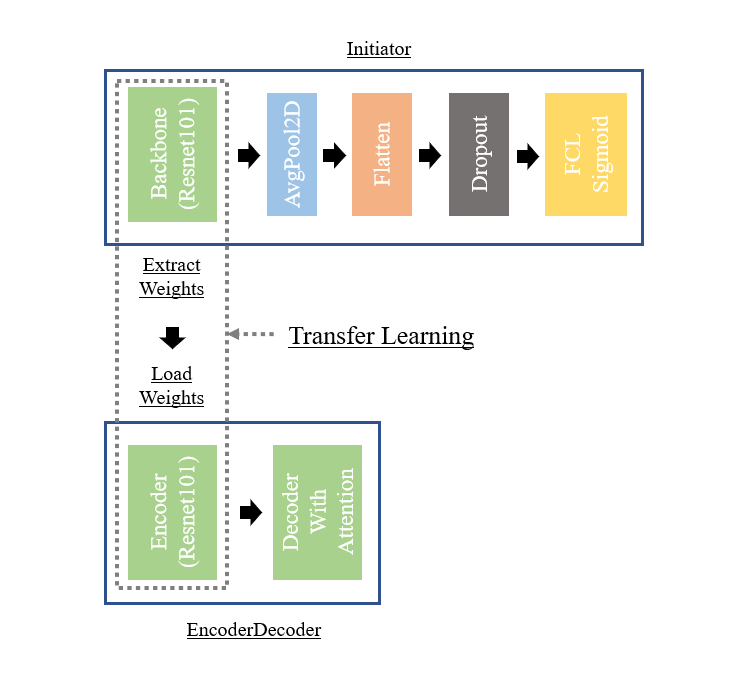
Illustration of the Transfer Learning part between the Initiator branch and the EncoderDecoder branch. The Initiator branch is voluntarily trained on a simplified problem in order to accustom the Resnet101 with images of molecules. Therefore, the Initiator branch is trained before the EncoderDecoder branch in order to extract the weights from the Resnet101 Backbone and load them on the Resnet101 Encoder of the EncoderDecoder branch.
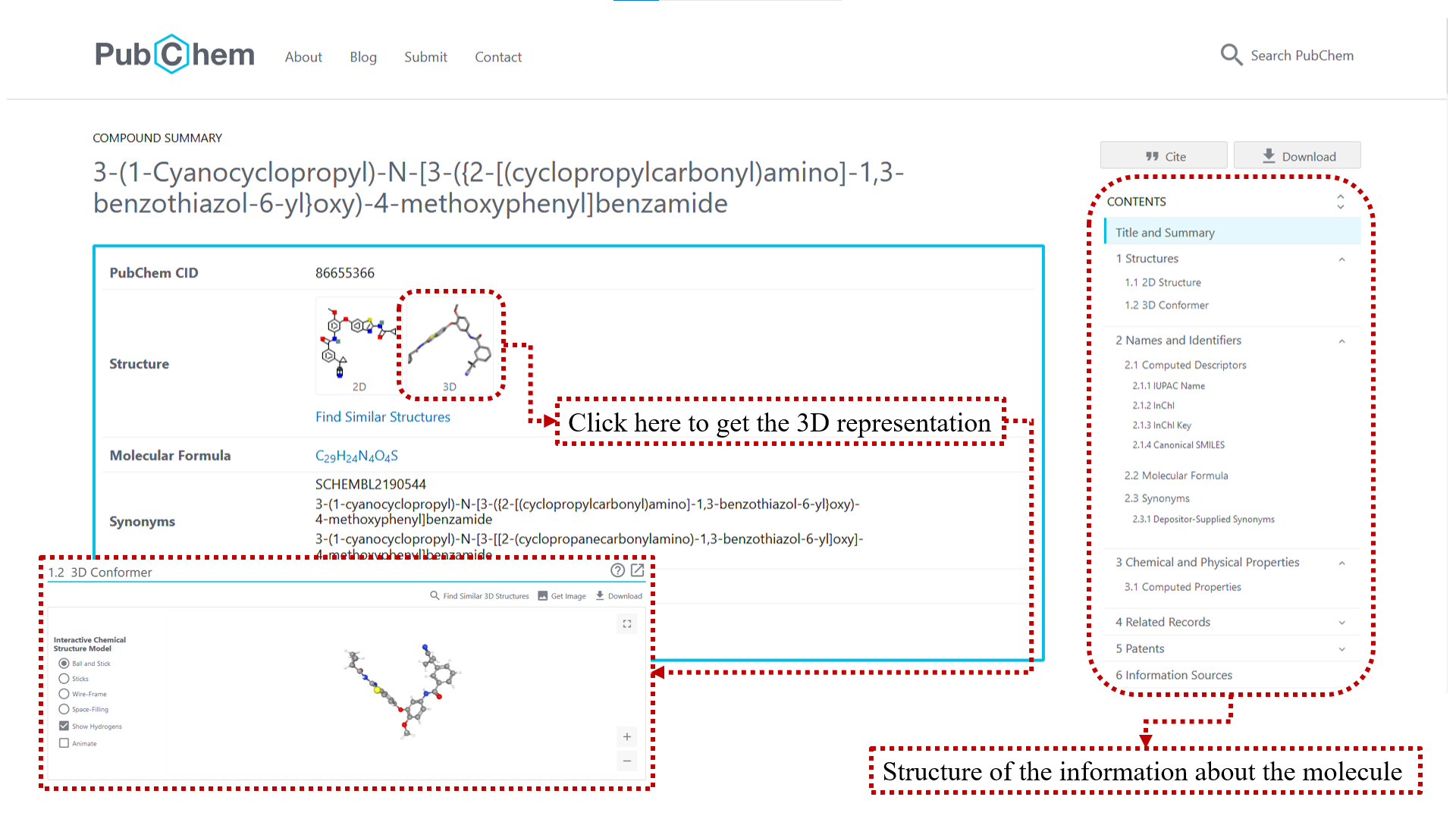
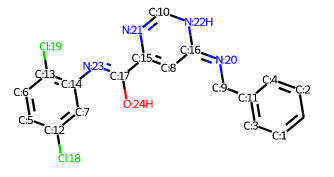
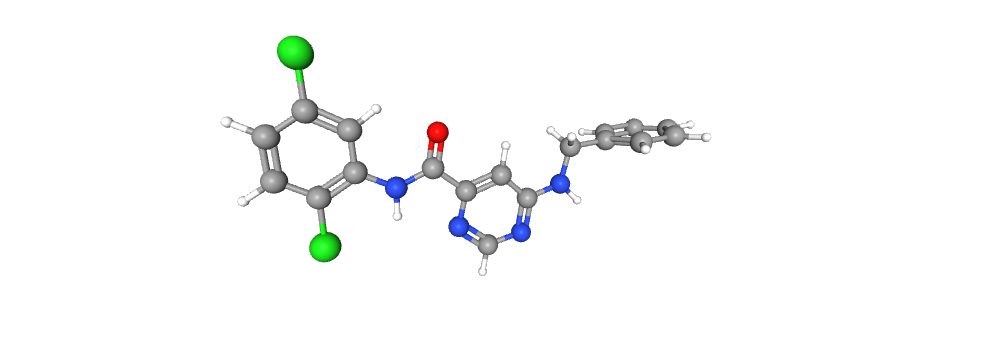
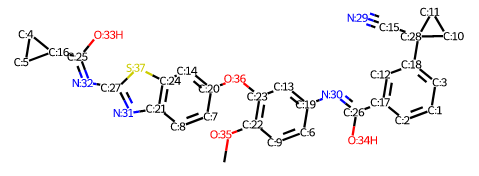
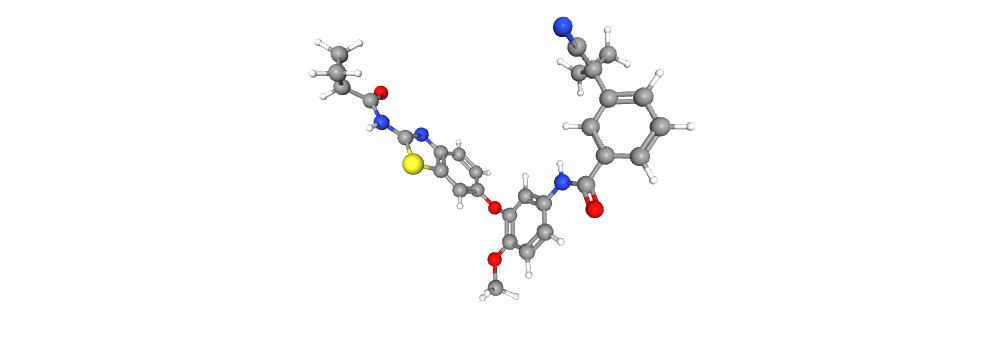
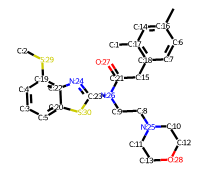
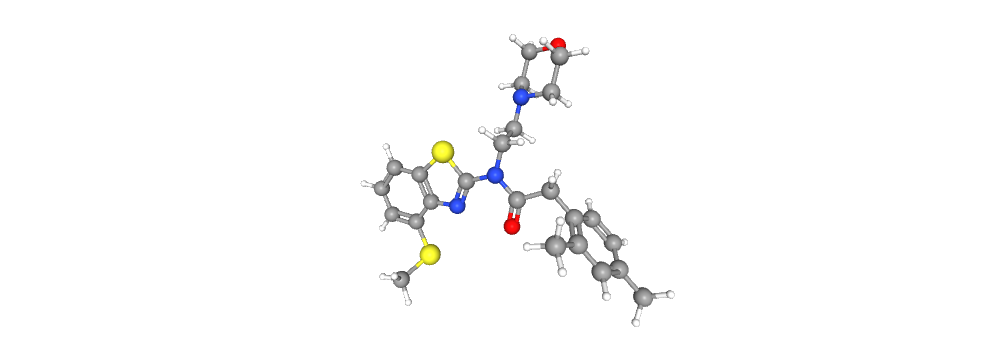
Representing the molecule in three dimensions allows to understand the structure of the molecule, interactions between bonds and atoms.
See Extras.ipynb (💥Notebook made with the Kaggle platform, package installations are different if you use Google Colab💥) to be able to plot molecules in 3D.
| 2D Structure | 3D Structure |
|---|---|
 |
 |
 |
 |
 |
 |
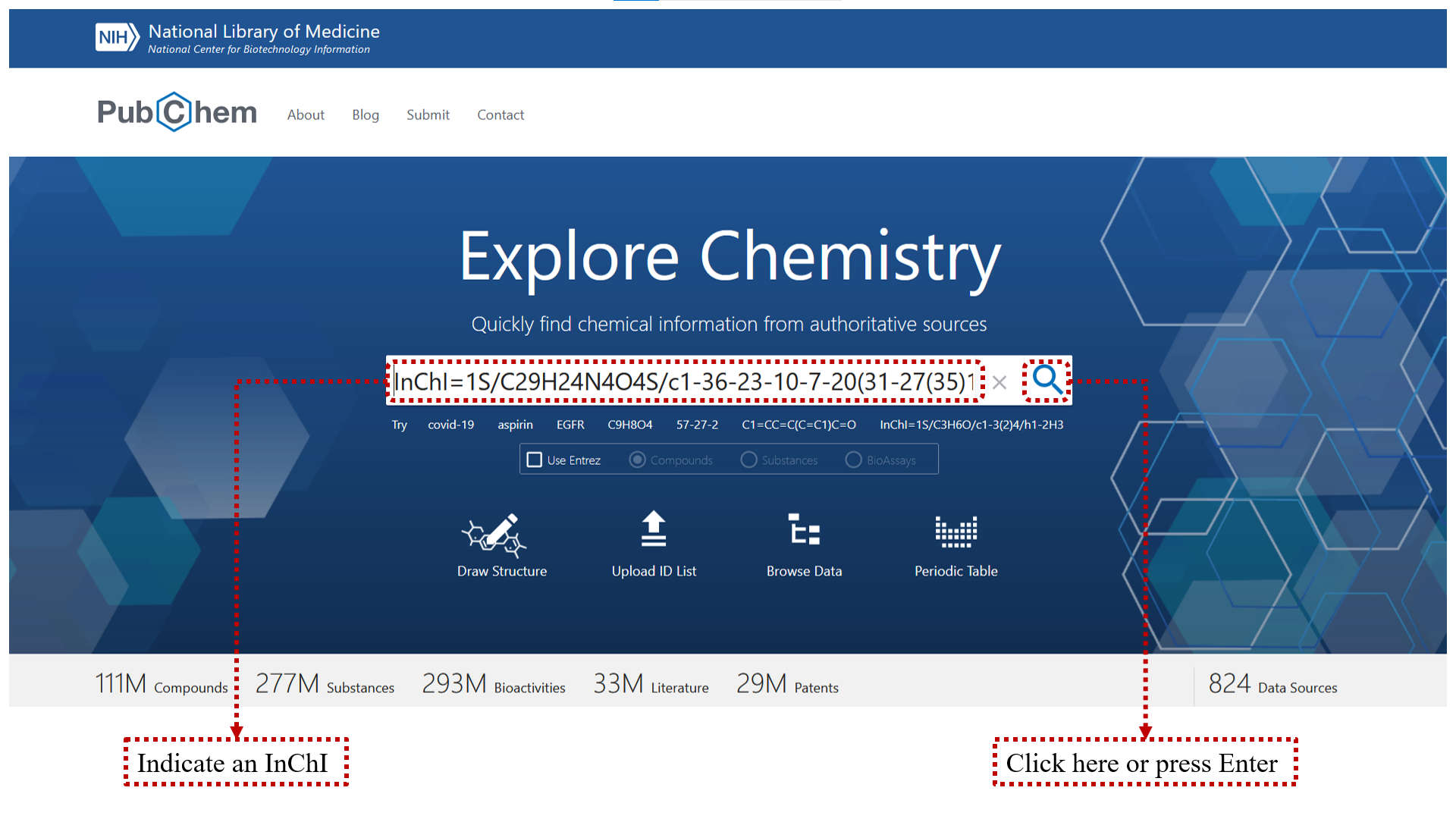
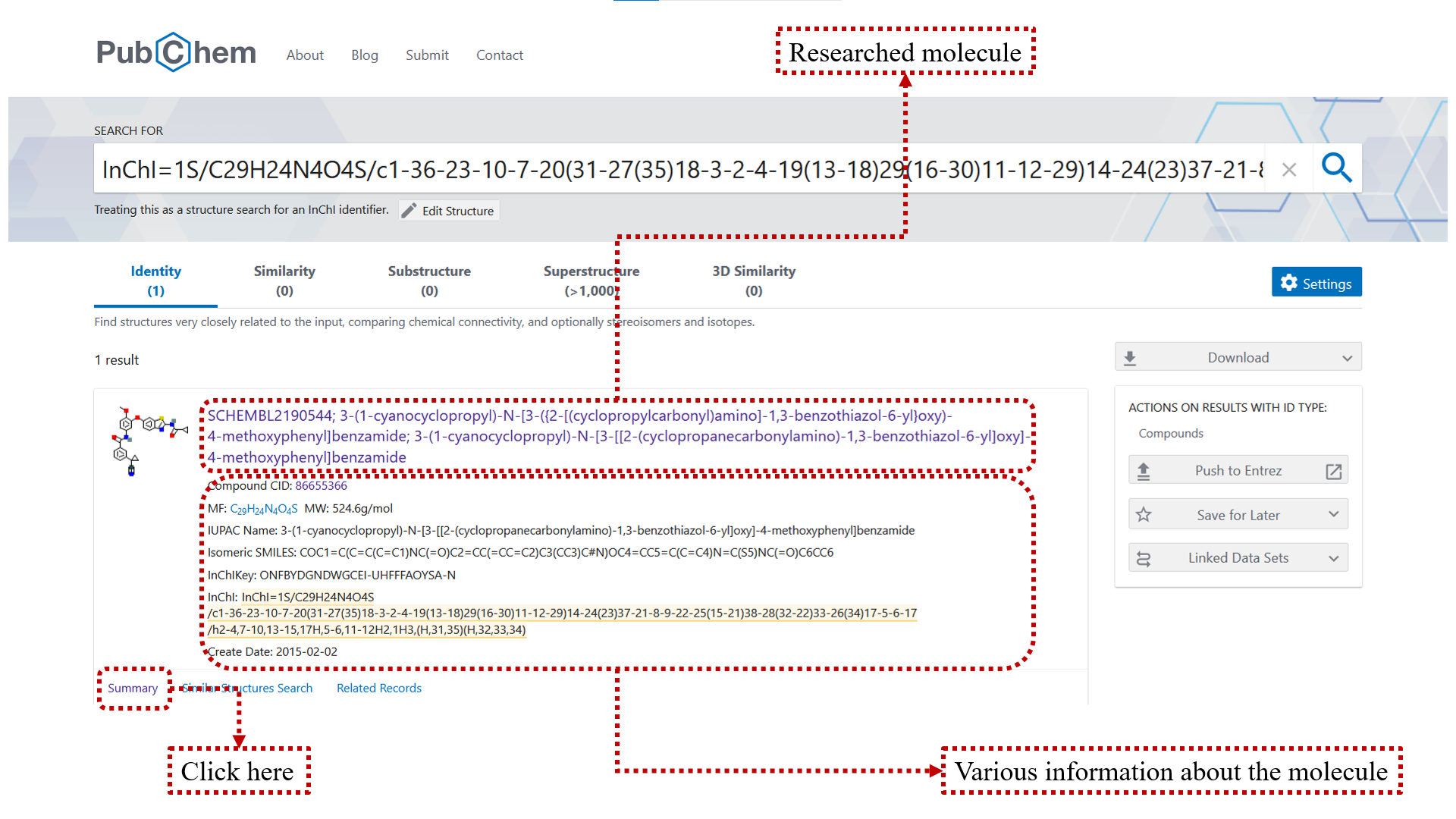
You can use PubChem.