- Author: Jan N Fuhg
- Organization: Cornell University
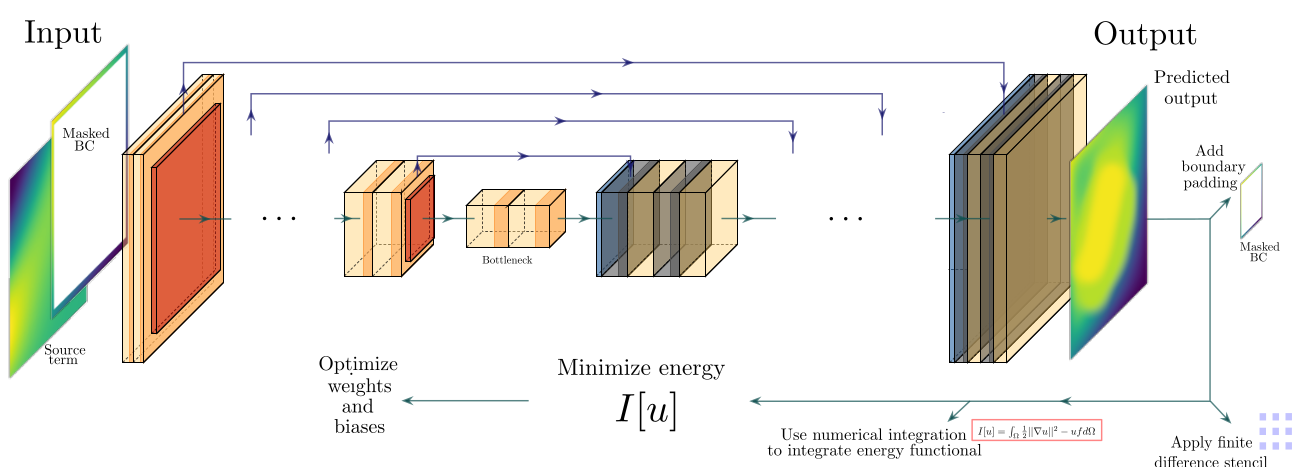
The parametric surrogate models for partial differential equations (PDEs) are a necessary component for many applications in computational sciences, and the convolutional neural networks (CNNs) have proven to be an excellent tool to generate these surrogates when parametric fields are present. CNNs are commonly trained on labeled data based on one-to-one sets of parameter-input and PDE-output fields. Recently, residual-based deep convolutional physics-informed neural network (DCPINN) solvers for parametric PDEs have been proposed to build surrogates without the need for labeled data. These allow for the generation of surrogates without an expensive offline-phase. In this work, we present an alternative formulation termed deep convolutional Ritz method (DCRM) as a parametric PDE solver. The approach is based on the minimization of energy functionals, which lowers the order of the differential operators compared to residualbased methods. Based on studies involving the Poisson equation with a spatially parameterized source term and boundary conditions, we find that CNNs trained on labeled data outperform DCPINNs in convergence speed and generalization abilities. The surrogates generated from the DCRM, however, converge significantly faster than their DCPINN counterparts, and prove to generalize faster and better than the surrogates obtained from both CNNs trained on labeled data and DCPINNs. This hints that the DCRM could make PDE solution surrogates trained without labeled data possible.
This code requires an Anaconda or Miniconda environment with a recent Python version. The complete repository can be cloned and installed locally. It is recommended to create a conda environment before installation. This can be done by the following the command line instructions
$ git clone https://github.com/FuhgJan/DCRM.git ./DCRM
$ cd DCRM
$ conda env create -f environment.yml
$ conda activate DCRM
$ python -m pip install . --user
Download the datasets from
Jan Fuhg. (2023). Poisson dataset for CNN training [Data set]. Zenodo. https://doi.org/10.5281/zenodo.8121086
and add them to the folder.
The provided example can then be run with
$ python main_DCRM.py
Outputs will be written to mixedDEM/outputs/vtk_files/ and can be opened with paraview.
Among others, the code requires the following packages as imports:
- NumPy for array handling
- Scipy for numerical solutions
- torch for the neural network and automatic differentiation libraries
- MatPlotLib for graphical output
If you use part of this code consider citing:
[1] Fuhg, J. N., et al. "Deep convolutional Ritz method: parametric PDE surrogates without labeled data." Applied Mathematics and Mechanics 44.7 (2023): 1151-1174.
This package comes with ABSOLUTELY NO WARRANTY. This is free software, and you are welcome to redistribute it under the conditions of the GNU General Public License (GPLv3)