A deep learning method to detect and measure partially occluded apples based on simultaneous modal and amodal instance segmentation
We provide a deep-learning method to better estimate the size of partially occluded apples. The method is based on ORCNN (https://github.com/waiyulam/ORCNN) and sizecnn (https://git.wur.nl/blok012/sizecnn), which extended Mask R-CNN network to simultaneously perform modal and amodal instance segmentation.
The amodal mask is used to estimate the fruit diameter in pixels, while the modal mask is used to measure in the depth map the distance between the detected fruit and the camera and calculate the fruit diameter in mm by applying the pinhole camera model.
See INSTALL.md
To use this method, please follow this procedure:
We have made our image-dataset (PApple_RGB-D-Size dataset) publicly available, which can be downloaded in the following link
It consists of 3925 RGB-D images of Fuji Apple Trees, including modal and amodal segmentation masks and diameter ground-truth of each annotated apple (15335 annotated apples).
In the following, we will assume that you have downloaded and extracted the datset in a folder named datasets, located in the base Amodal_Fruit_Sizing folder:
mkdir datasets
cd datasets
# if you do not have wget, you can download the file by any other method.
wget https://zenodo.org/record/7260694/files/PApple_RGB-D-Size_dataset_v4.zip
unzip PApple_RGB-D-Size_dataset_v4.zip
rm PApple_RGB-D-Size_dataset_v4.zip
cd ..
Execute Train_AmodalFruitSize.py to train the network:
python trainval_net.py \
--num_iterations $MAX_NUM_OF_ITERATIONS --checkpoint_period $CHECKPOINT_PERIOD \
--eval_period $EVAL_PERIOD --batch_size $BATCH_SIZE \
--learing_rate $LEARNING_RATE --LR_decay $WEIGHT_DECAY \
--experiment_name $OUTPUT_FOLDER_NAME --dataset_path $DATASET_DIRECTORY \
--batch_size_per_image $BATCH_SIZE_PER_IMAGE --output_dir=$OUTPUT_DIRECTORY'
example:
python Train_AmodalFruitSize.py --num_iterations 4000 --checkpoint_period 500 --eval_period 500 --batch_size 4 --learing_rate 0.02 --LR_decay 0.0001 --experiment_name "trial01" --dataset_path "./datasets/data/" --output_dir='./output/' --batch_size_per_image 512
If you want to evaluate the detection performance of a pre-trained model on our set execute AmodalFruitSize_val_test_evaluation.py to test the network:
python AmodalFruitSize_val_test_evaluation.py \
--experiment_name $EXPERIMENT_NAME --test_name $TEST_ID_NAME \
--dataset_path $DATASET_DIRECTORY --split $DATASET_SPLIT \
--output_dir $OUTPUT_DIRECTORY \
--weights $WEIGHTS_FILE --focal_length $CAMERA_FOCAL_LENGTH \
--iou_thr $INTESECTION_OVER_UNION --nms_thr $NON_MAXIMUM_SUPRESION \
--confs $MINIMUM_CONFIDENCES_TO_TEST
example:
python AmodalFruitSize_val_test_evaluation.py --experiment_name 'trial01' --test_name 'eval_01' --dataset_path './datasets/data/' --split 'test' --output_dir='./output/' --weights './output/trial01/model_0002999.pth' --focal_length 5805.34 --iou_thr 0.5 --nms_thr 0.1 --confs '0.0,0.05,0.1,0.15,0.2,0.25,0.3,0.35,0.4,0.45,0.5,0.55,0.6,0.65,0.7,0.75,0.8,0.85,0.9,0.95,0.99'
If you want to run the detection and sizing method on your own images with a pre-trained model, download the pretrained model or train your own models at first, and then execute AmodalFruitSize_inference.py:
python AmodalFruitSize_inference.py \
--experiment_name $OUTPUT_FOLDER_NAME --dataset_path $DEMO_DATA_DIRECTORY \
--output_dir $OUTPUT_DIRECTORY \
--test_name $FOLDER_FROM_THE_DATASET_CONTAINING_THE_IMAGES --weights $WEIGHTS_FILE \
--nms_thr $NON_MAXIMUM_SUPRESION --confs $MINIMUM_CONFIDENCES_TO_TEST
example:
python AmodalFruitSize_inference.py --experiment_name 'demo' --dataset_path "./demo/" --test_name 'demo_data' --output_dir='./output/' --weights './output/trial01/model_0002999.pth' --nms_thr 0.1 --conf 0
Note: To run the AmodalFruitSize_inference.py code the reader can use the images provided in './demo/demo_data'. The pre-trained weights can be downloaded in the following link. It is also possible to run inference on the test/val/train splits of the downloaded dataset:
python AmodalFruitSize_inference.py --experiment_name 'test_set' --dataset_path "./datasets/" --test_name 'data' --output_dir='./output/' --weights './output/trial01/model_0002999.pth' --nms_thr 0.1 --conf 0 --split='test'
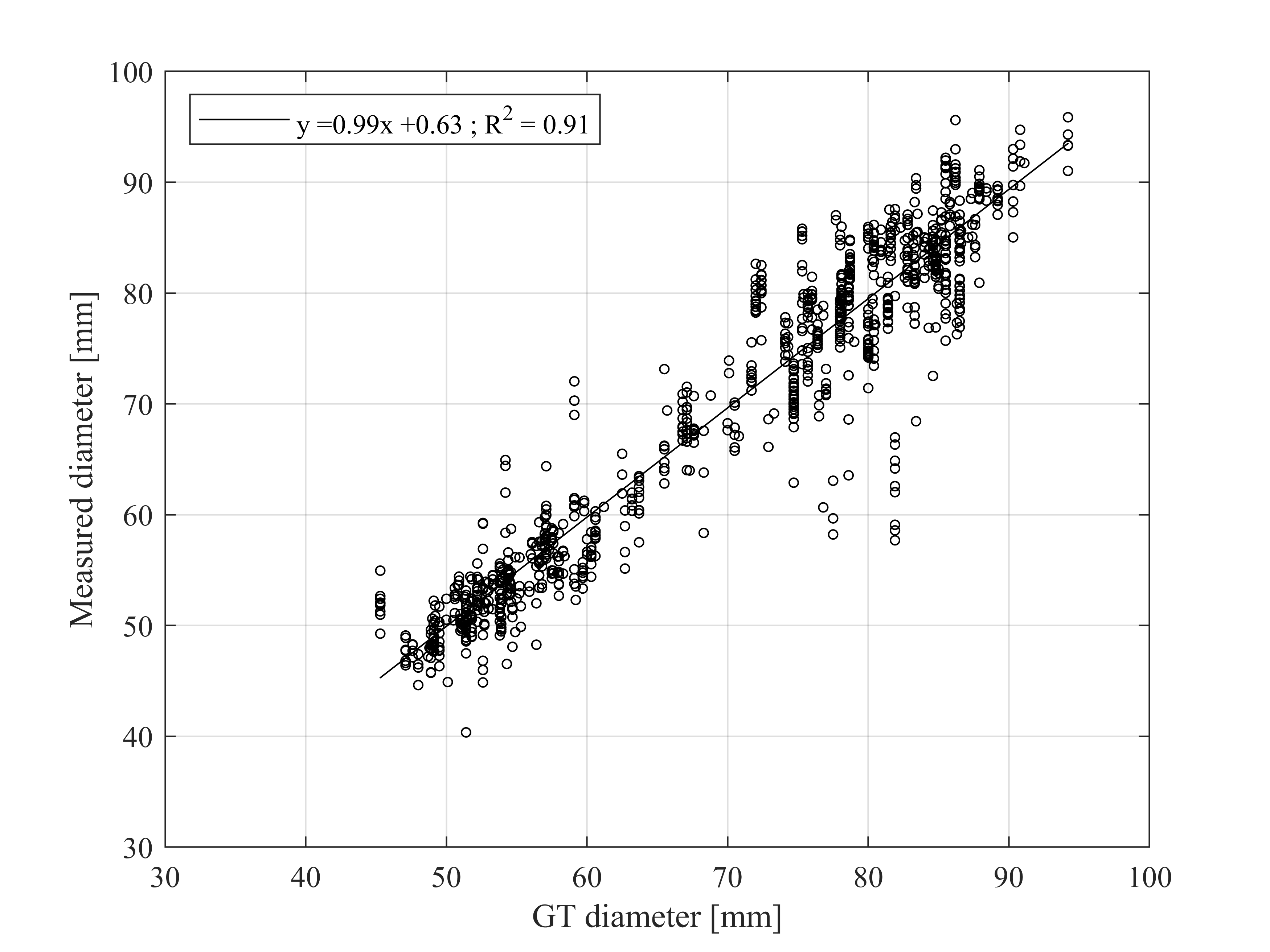
Since the percentage of visibility can be automatically estimated by using modal and amodal masks, we evaluated the sizing performance at different fruit visibility levels:
Results when measuring apples automatically detected with a visibility higher than 60%:
| Green apples | Ripe apples | Green + Ripe apples | |
|---|---|---|---|
| MAE (mm) | 2.05 | 3.34 | 2.93 |
| MBE (mm) | -0.54 | -0.02 | -0.19 |
| MAPE (%) | 3.79 | 4.27 | 4.19 |
| RMSE (mm) | 2.80 | 4.59 | 4.14 |
Linear correlation between the ground truth (GT) diameter and the automatically measured diameter of apples detected in the test set with a visibility higher than 60%:
Qualitative results:
Green values correspond to the ground truth and white values are the measures automatically estimated with the present method. Only the apples with an estimated visibility higher than 60% where measured.

| Backbone | Dataset | Weights |
|---|---|---|
| ResNext_101_32x8d_FPN_3x | PApple_RGB-D-Size | model_0002999.pth |
Our software was forked from sizecnn (https://git.wur.nl/blok012/sizecnn), which was forked from ORCNN (https://github.com/waiyulam/ORCNN), which was forked from Detectron2 (https://github.com/facebookresearch/detectron2). As such, our CNN's will be released under the Apache 2.0 license.
Please cite our research article or dataset when using our software and/or dataset:
@article{XXXXX2022,
title = {Looking Behind Occlusions Through Amodal Segmentation for Robust On-tree Fruit Size Measurement},
author = {Jordi Gené-Mola and Mar Ferrer-Ferrer and Eduard Gregorio and Pieter M. Blok and Jochen Hemming and Josep-Ramon Morros and Joan R. Rosell-Polo and Verónica Vilaplana and Javier Ruiz-Hidalgo},
journal = {Computers and Electronics in Agriculture},
volume = {209},
pages = {107854},
year = {2023},
doi = {https://doi.org/10.1016/j.compag.2023.107854}
}