TotalSegmentator packaged as an AIDE Application, based on
the open-source
MONAI Application Package (MAP) standard.
View repo
·
Report Bug
·
Request Feature
The input to the TotalSegmentator MAP is a folder containing CT DICOM .dcm files. The output is an RT Struct DICOM .dcm
file containing the contours of all the segmented regions.
The TotalSegmentator MAP consists of three operators:
dcm2nii_operator.py– converts the input CT DICOM files to NIfTI format required by TotalSegmentator, using dcm2niixtotalsegmentator_operator.py– runs TotalSegmentatorrtstructwriter_operator.py– converts the output segmentations from NIfTI to DICOM RT Struct format, using rt-utilsclinrev_pdf_operator.py– creates a DICOM encapsulated PDF showing three views of the input CT dataset overlaid with the segmentations generated by TotalSegmentator
- GPU-enabled machine
- Docker
- Required for running the MAP. Not required for running source code
- Download
git clone https://github.com/GSTT-CSC/TotalSegmentator-AIDE.git- Setup virtual env
cd TotalSegmentator-AIDE
python -m venv venv
source venv/bin/activate
pip install --upgrade pip setuptools wheel
pip install -r requirements.txt- Create
inputandoutputdirectories
mkdir input output- Ensure Python venv running
- Copy DICOM
.dcmfiles toinput/directory
monai-deploy exec app -i input/ -o -output/
# alternatively:
python app -i input/ -o output/- Ensure Python venv running
- Ensure Docker running
- Copy DICOM
.dcmfiles toinput/directory
Important: we create an initial MAP map-init upon which we build any 3rd-party non-Python software (e.g. dcm2niix).
The final MAP is called map
# Initial packaging of MAP
monai-deploy package app --tag ghcr.io/gstt-csc/totalsegmentator-aide/map-init:0.2.0 -l DEBUG
# Build 3rd-party software on top of MAP
docker build -t ghcr.io/gstt-csc/totalsegmentator-aide/map:0.2.0 app/
# Test MAP with MONAI Deploy
monai-deploy run ghcr.io/gstt-csc/totalsegmentator-aide/map:0.2.0 input/ output/
# Push initial MAP and final MAP to GHCR
docker push ghcr.io/gstt-csc/totalsegmentator-aide/map-init:0.2.0
docker push ghcr.io/gstt-csc/totalsegmentator-aide/map:0.2.0Enter Docker container for testing
docker run --gpus all -it --rm -v /TotalSegmentator-AIDE/input:/var/monai/input/ --entrypoint /bin/bash ghcr.io/gstt-csc/totalsegmentator-aide/map:0.2.0Run on specified GPU if machine has >1 available
CUDA_VISIBLE_DEVICES=2 monai-deploy run ghcr.io/gstt-csc/totalsegmentator-aide/map:0.2.0 input/ output/Reminder: if GHCR requires Personal Access Token
echo $CR_PAT | docker login ghcr.io -u <github-username> --password-stdin
docker push ghcr.io/gstt-csc/totalsegmentator-aide/map-init:0.2.0Note: two containers are present under GitHub Packages:
The map version is for general usage and deployment. The map-init version is for developers who wish to build the
MAP from scratch as part of the two-step build process described above.
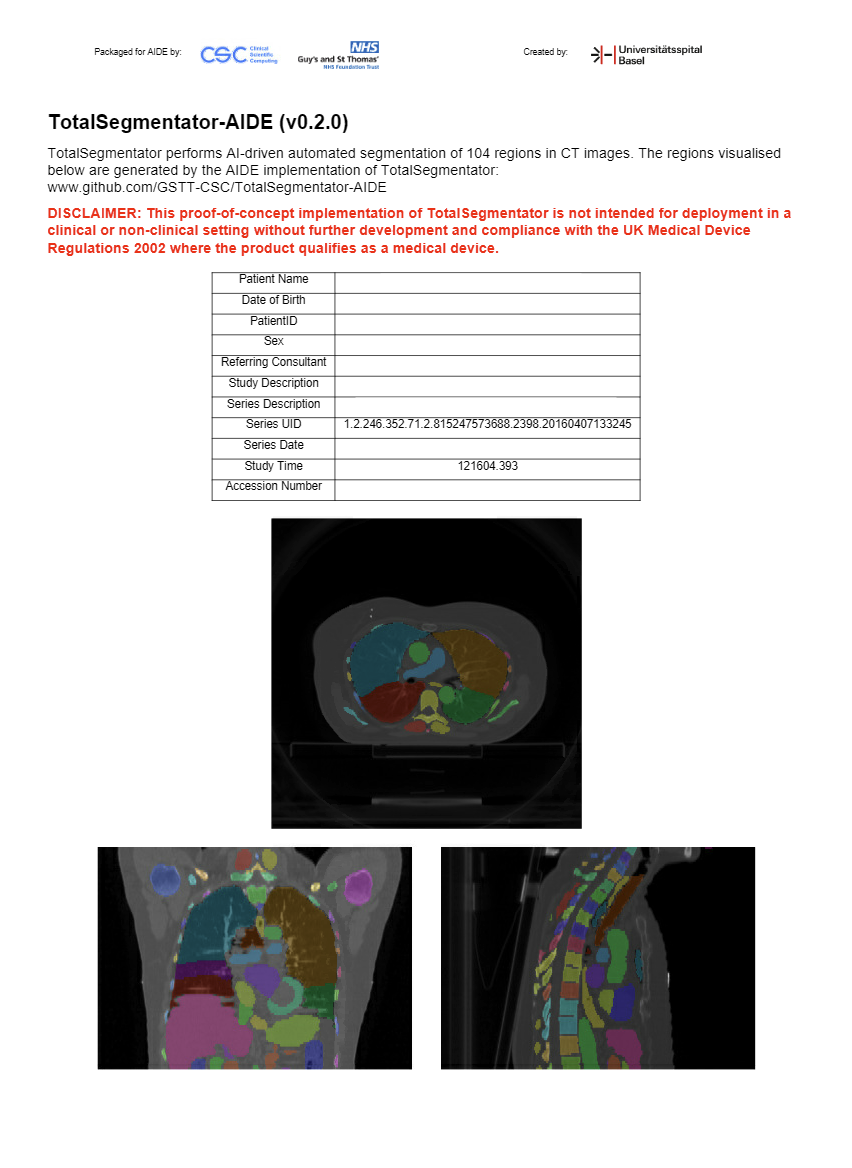
AIDE Clinical Review is a user interface within AIDE for assessing the quality of AI results. It can display DICOM
Encapsulated PDFs. The screenshot below is an example PDF generated by clinrev_pdf_operator.py:
To run TotalSegmentator on AIDE, two files from the app/workflows directory are required, namely:
totalsegmentator.json– AIDE Clinical Workflow file.- Effectively a sequence of tasks run on AIDE
totalsegmentator-argo-template.yaml– Argo Workflow Template file.- Called by the AIDE Clinical Workflow file. Executes the Totalsegmentator-AIDE MAP using Argo. This is the central task within the AIDE Clinical Workflow file.
The app/workflows directory also contains four illustrative Argo Workflow Templates depending on system performance.
low, medium, high and dgx examples are provided with different CPU and RAM settings.
pycairo
If you encounter an issue during installation where pycairo cannot be installed, you may need to install libcairo2-dev via the following (assuming Linux):
sudo apt install libcairo2-dev