PlanText: Gradually Masked Guidance to Align Image Phenotype with Trait Description for Plant Disease Texts Tool
We propose a new approach to enhance plant disease diagnosis using multimodal artificial intelligence techniques. progressive masking of disease image diagnostic text, we aim to more accurately align plant phenotypes with trait descriptions. Our approach consists of three stages: extracting structural features of plant shape descriptions and integrating reconstructed utterances into phenotype descriptions. integrating the reconstructed statements into the phenotypic descriptions, and aligning image-text features to generate dialog descriptions. The experimental results show superior performance compared to existing models, including outperformance of GPT-4 and GPT-4o. Future work includes expanding the dataset and improving feature selection and alignment to improve accuracy.
clone this repository,and build it with the following command.
cd PlanText
pip install -r requirements.txtThe dataset used by our human annotation, You can get it by visiting our website.
The framework of the proposed method
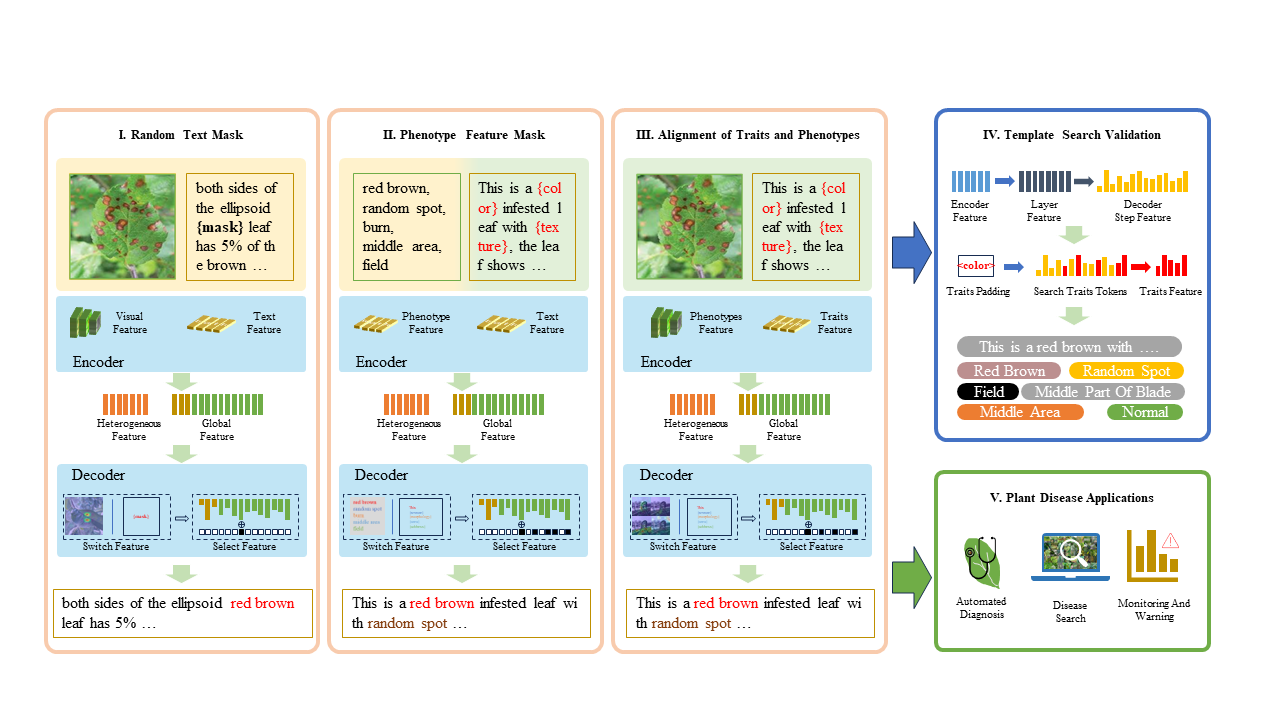
A way to incrementally train PlanText models on your dataset:
python -u main.py \
--model ViTGPT2-Our \
--model_path=output18 \
--batch_size 32 \
--pre_epoch 40 \
--annotation your_manually_labeled_Dir
--image_path you_images_base_path
or edit train debug demo train/train.sh
sh train/train.shYou can generate text and labels with --template_search. But batch_size must be 1
python -u main2.py --batch_size 1 --template_search --test_epoch 20 --model_path=output18 --batch_size 1
Or use --test to generate all the required data.
python -u main2.py --batch_size 1 --test --test_epoch 20 --model_path=output18 --batch_size 1
--test_epoch is to specify the model batch --model_path is model save path.
Part of our code comes from the following work, and we are very grateful for their contributions to relevant research:
transformers,
AdaptiveAttention.
We thank the authors and appreciate their great works!