This deep learning dataset was developed for research on biologically plausible error-backpropagation and deep learning in spiking neural networks. It serves as an alternative to e.g. the MNIST dataset providing the following advantages:
- Very clear gap between the accuracies reached by a linear classifier/shallow network, a deep neural network and a deep network with frozen lower weights
- Smaller and therefore faster to train
- Symmetric input design allows successful training of neuron models without intrinsic bias
For more information see:
The Yin-Yang dataset; L. Kriener, J. Göltz, M. A. Petrovici; https://doi.org/10.1145/3517343.3517380 (https://arxiv.org/abs/2102.08211)
As shown in the image below each data sample consists of 4 values (x1, y1, x2, y2) where x2 and y2 are generated by a mirrored axis. This symmetry is helpful when training neuron models without intrinsic bias.
Each data sample is classified into one of the three classes yin (blue), yang (orange) and dot (green). All classes have approximately the same amount of samples.
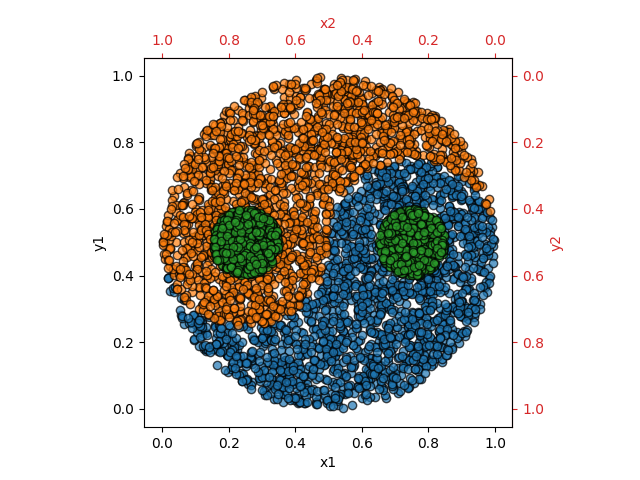
In order to produce a balanced dataset, the data is generated with rejection sampling:
- draw random integer between 0 and 2, this is the desired class
cof the next sample that will be generated - draw
x1andy1value and calculate the correspondingx2andy2 - check if the drawn sample is of class
c- if yes: keep sample
- if no: draw new
x1andy1and check again
- repeat until chosen number of samples are produced
The equations for the calclulation of the Yin-Yang shape are inspired by: https://link.springer.com/content/pdf/10.1007/11564126_19.pdf (Appendix A).
This dataset is designed to make use of the PyTorch dataset and dataloader architecture, but (with some minor changes) it can also be used in any other deep learning framework.
A detailed example is shown in example.ipynb.
Initializing the dataset:
from dataset import YinYangDataset
dataset_train = YinYangDataset(size=5000, seed=42)
dataset_validation = YinYangDataset(size=1000, seed=41)
dataset_test = YinYangDataset(size=1000, seed=40)Note: It is very important to give different seeds for trainings-, validation- and test set, as the data is generated randomly using rejection sampling. Therefore giving the same seed value will result in having the same samples in the different datasets!
Setting up PyTorch Dataloaders:
from torch.utils.data import DataLoader
batchsize_train = 20
batchsize_eval = len(dataset_test)
train_loader = DataLoader(dataset_train, batch_size=batchsize_train, shuffle=True)
val_loader = DataLoader(dataset_validation, batch_size=batchsize_eval, shuffle=True)
test_loader = DataLoader(dataset_test, batch_size=batchsize_eval, shuffle=False)Even though this dataset is designed for PyTorch, the following easy code changes make it useable in any deep learning framework:
- In
dataset.pydelete line 2
from torch.utils.data.dataset import Dataset- and replace in line 5
class YinYangDataset(Dataset):by
class YinYangDataset(object):The dataset can now be loaded as shown above. The data can be iterated over, saved to file or imported in other frameworks.
These references are generated with the same settings for network layout and training (hidden layer of 30 neurons for the ANN, batchsize of 20, learning rate of 0.01, adam optimizer, 300 epochs for the ANN as well as the shallow network) as in example.ipynb. The training was repeated 20 times with randomly chosen weight initializations. The accuracies are given as mean and standard devaition over the 20 runs.
For more details see https://doi.org/10.1145/3517343.3517380 (https://arxiv.org/abs/2102.08211).
| Network | Test accuracy [%] |
|---|---|
| ANN (4-30-3) | 97.6 +- 1.5 |
| ANN (4-30-3) frozen lower weights | 85.5 +- 5.8 |
| Shallow network | 63.8 +- 1.0 |
Up until now this dataset has been used in the following publications:
- Fast and deep: energy-efficient neuromorphic learning with first-spike times; J. Göltz∗, L. Kriener∗, A. Baumbach, S. Billaudelle, O. Breitwieser, B. Cramer, D. Dold, A. F. Kungl, W. Senn, J. Schemmel, K. Meier, M. A. Petrovici; https://www.nature.com/articles/s42256-021-00388-x (https://arxiv.org/abs/1912.11443)
- EventProp: Backpropagation for Exact Gradients in Spiking Neural Networks; T. Wunderlich and C. Pehle; https://www.nature.com/articles/s41598-021-91786-z (https://arxiv.org/abs/2009.08378)
- A scalable approach to modeling on accelerated neuromorphic hardware; E. Müller, E. Arnold, O. Breitwieser, M. Czierlinski, A. Emmel, J. Kaiser, C. Mauch, S. Schmitt, P. Spilger, R. Stock, Y. Stradmann, J. Weis, A. Baumbach, S. Billaudelle, B. Cramer, F. Ebert, J. Göltz, J. Ilmberger, V. Karasenko, M. Kleider, A. Leibfried, C. Pehle, J. Schemmel; https://www.frontiersin.org/articles/10.3389/fnins.2022.884128/full
- Exact gradient computation for spiking neural networks through forward propagation; J. H. Lee, S. Haghighatshoar, A. Karbasi; https://proceedings.mlr.press/v206/lee23b.html (https://arxiv.org/abs/2210.15415)
- Learning efficient backprojections across cortical hierarchies in real time; K. Max, L. Kriener, G. P. García, T. Nowotny, W. Senn, M. A. Petrovici; https://arxiv.org/abs/2212.10249
- hxtorch.snn: Machine-learning-inspired Spiking Neural Network Modeling on BrainScaleS-2; P. Spilger, E. Arnold, L. Blessing, C. Mauch, C. Pehle, E. Müller, J. Schemmel; https://dl.acm.org/doi/abs/10.1145/3584954.3584993
- Safe semi-supervised learning for pattern classification; J. Ma, G. Yu, W. Xiong, X. Zhu; https://doi.org/10.1016/j.engappai.2023.106021
- Event-based Backpropagation for Analog Neuromorphic Hardware; C. Pehle, L. Blessing, E. Arnold, E. Müller, J. Schemmel; https://arxiv.org/abs/2302.07141
If you would like to use this dataset in a publication feel free to do so and please contact me so that this list can be updated (or update it yourself via a pull request).
The data used in the publication "Fast and deep: energy-efficient neuromorphic learning with first-spike times" is also uploaded as .npy files in publication_data/.