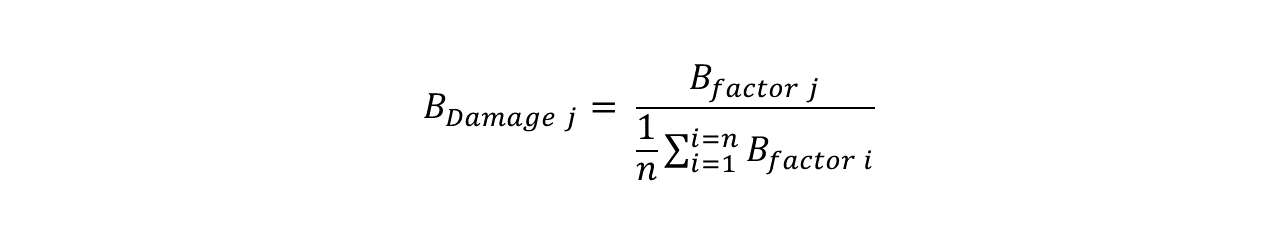A program to calculate the BDamage , Bnet and Bnet-percentile metrics to quantify the extent of specific radiation damage present within a cryo-temperature protein crystal (PX) structure. RABDAM will run on any standard format PDB or mmCIF file, but since it assesses radiation damage via calculating the above B-factor-based metrics, it should only be run towards the end of refinement/on models deposited in the PDB.
RABDAM is a command line program. To run the program with its recommended default parameter values, execute:
python rabdam.py –f XXXX
, where XXXX is the 4 character PDB accession code of the MX structure under study. Alternatively, you can provide RABDAM with a file path to a locally saved PDB file:
python rabdam.py –f /path/to/pdb_file.pdb
, or to a locally saved mmCIF file:
python rabdam.py –f /path/to/mmcif_file.cif
The Bnet and Bnet-percentile metrics should only by calculated for cryo-temperature protein crystal structures (the structure may contain other components such as nucleic acids, but it must contain protein).
See the “Usage” section below for further details.
We have released RABDAM version 2. The main changes from version 1 are:
- RABDAM version 2 now calculates Bnet-percentile values
- We have introduced the
filterflag to allow users to check their input models meet the requirements for Bnet and Bnet-percentile calculation set out in Shelley & Garman, 2022. - We have switched over from using PDBCUR to using cctbx to generate a copy of the unit cell. Due to small rounding differences in the xyz coordinates output by the two programs, occasionally this affects the BDamage packing density calculation, and so for some models there are slight differences in the Bnet values output from RABDAM version 2 as compared to RABDAM version 1.
During macromolecular crystallography (MX) data collection, X-rays are also absorbed by and deposit energy within the crystal under study, causing damage. This damage can result in localised chemical changes to the crystalline macromolecule copies, such as disulfide bond cleavage, and the decarboxylation of glutamate and aspartate side chains. Such specific radiation damage manifestations can lead to incorrect biological conclusions being drawn from an MX structure if they are not identified and accounted for.
The chemical changes induced by specific radiation damage cause an accompanying increase in the atomic B-factor values of affected sites. However, multiple factors can affect an atom’s B-factor value in addition to radiation damage, the most important of these being its mobility. The increase in atomic B-factor caused by specific radiation damage is insufficiently large to distinguish damage from mobility.
There is a strong positive correlation between the mobility of an atom within a crystal structure and its packing density, i.e. the number of atoms present in its local environment. The BDamage metric is full isotropic atomic B-factor corrected for packing density: specifically, the BDamage value of an atom j is equal to the ratio of its B-factor to the average B-factor of atoms 1 to n which occupy a similar packing density environment to atom j. The BDamage metric has been shown to identify expected sites of specific radiation damage in damaged MX structures (Gerstel et al., 2015).
The method of calculating an atom’s BDamage value is summarised in the diagram below:
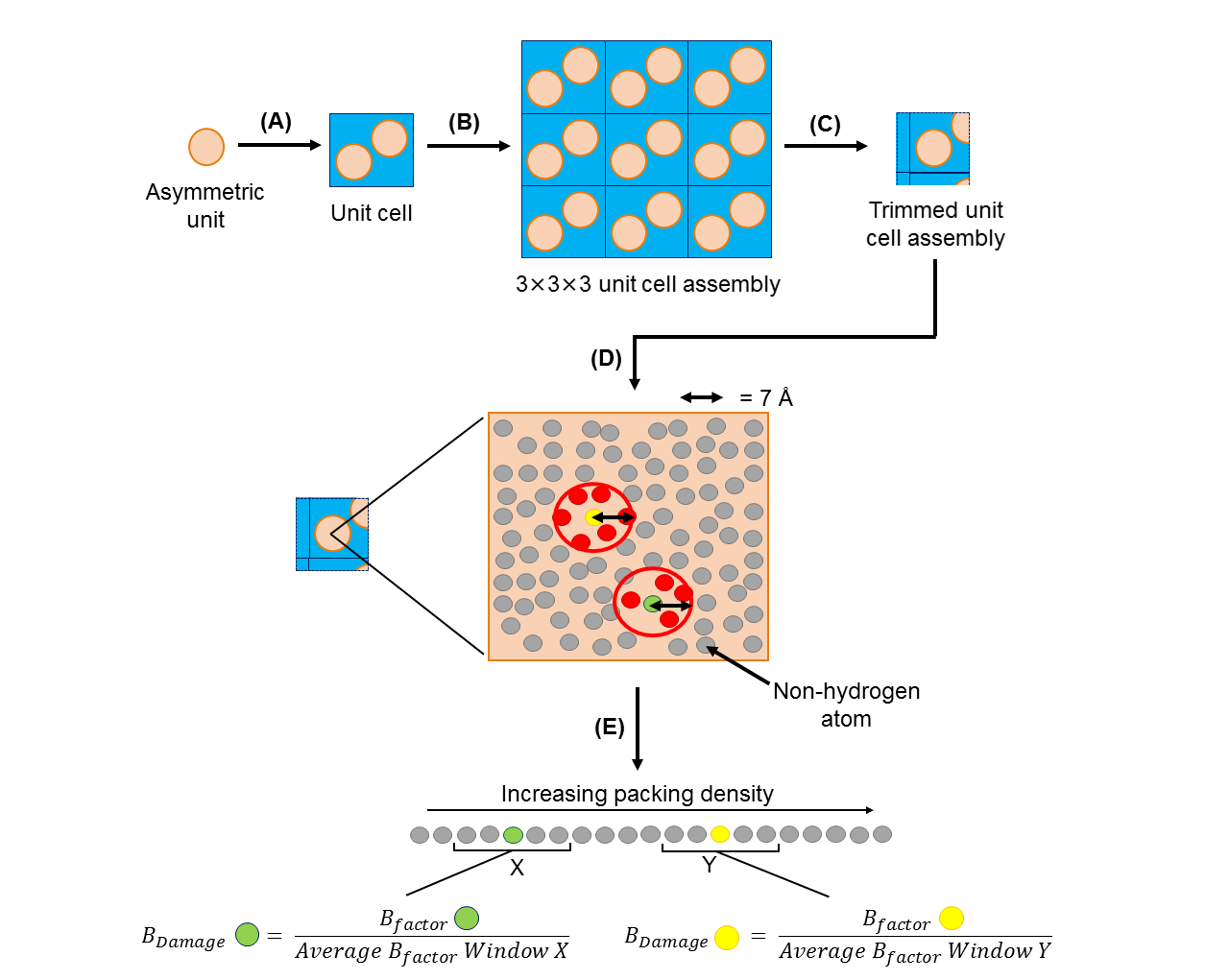 Schematic illustrating the calculation of the BDamage metric. From an input PDB/mmCIF file of the asymmetric unit of a protein crystal structure of interest, RABDAM (A) generates a copy of the unit cell, followed by (B) a 3x3x3 assembly of unit cells. (C) Atoms in the 3x3x3 unit cell assembly that lie further than 7 Å from the asymmetric unit are discounted. (D) The packing density of an atom j in the asymmetric unit is calculated as the number of atoms within a 7 Å radius. (E) Asymmetric unit atoms are ordered by packing density; the BDamage value of atom j is then calculated as the ratio of its B-factor to the average of the B-factor values of atoms grouped, via a sliding window, as occupying a similar packing density environment. Note that hydrogen atoms are not considered in the calculation of BDamage.
Schematic illustrating the calculation of the BDamage metric. From an input PDB/mmCIF file of the asymmetric unit of a protein crystal structure of interest, RABDAM (A) generates a copy of the unit cell, followed by (B) a 3x3x3 assembly of unit cells. (C) Atoms in the 3x3x3 unit cell assembly that lie further than 7 Å from the asymmetric unit are discounted. (D) The packing density of an atom j in the asymmetric unit is calculated as the number of atoms within a 7 Å radius. (E) Asymmetric unit atoms are ordered by packing density; the BDamage value of atom j is then calculated as the ratio of its B-factor to the average of the B-factor values of atoms grouped, via a sliding window, as occupying a similar packing density environment. Note that hydrogen atoms are not considered in the calculation of BDamage.
The Bnet metric is a derivative of the (per-atom) BDamage metric: Bnet summarises in a single value the total extent of specific radiation damage suffered by a cryo-temperature protein crystal structure. One of the best-characterised chemical changes resulting from specific radiation damage that occurs at cryo-temperatures within proteins is the decarboxylation of Glu and Asp residues: the Bnet metric is calculated from a kernel density estimate of the BDamage values of a structure’s Glu and Asp side chain oxygen atoms as the ratio of the area under the curve either side of the median of the (overall) BDamage distribution.
Bnet is weakly correlated with resolution. To obtain a metric that is not correlated with resolution, Bnet-percentile is calculated as the percentile ranking of a model’s Bnet value within the subset of models derived from cryo-datasets which are closest in resolution (the 1000 structures closest in resolution in the PDB are first identified, and subsequently all structures falling within this resolution range are included in the percentile calculation). Since Bnet-percentile is not correlated with resolution, it is the recommended metric for damage comparison between structures.
The method of calculating the Bnet value for a protein structure is summarised in the diagram below:
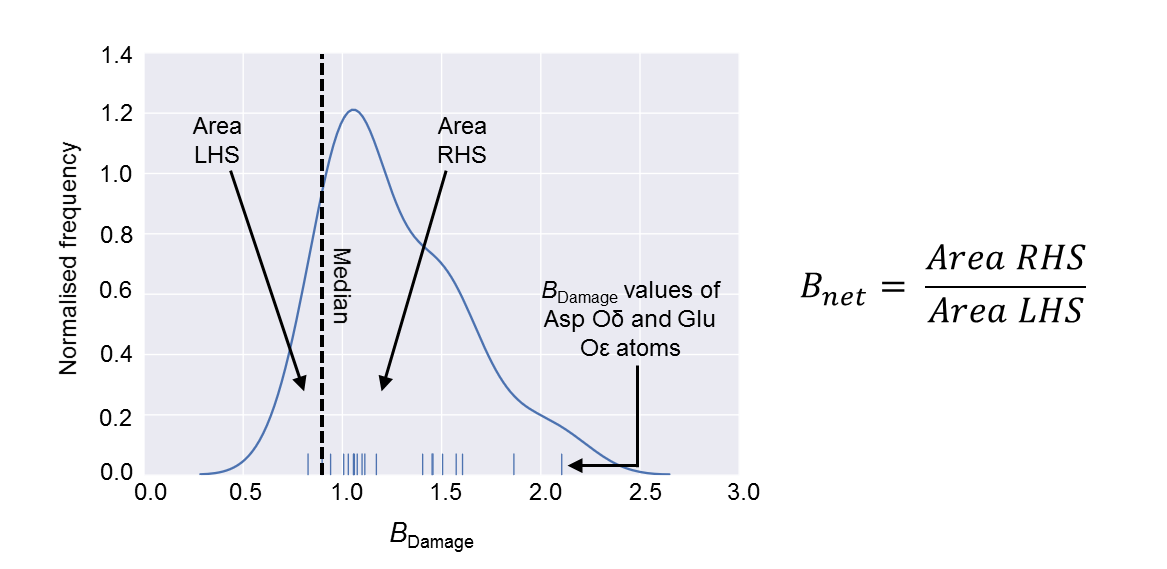 The Bnet metric is calculated by first plotting a kernel density estimate of the BDamage values of Glu and Asp side chain oxygen atoms. The area under the curve to the left- (LHS) and right-hand side (RHS) of the median BDamage value of all atoms in the structure is then measured, and Bnet is calculated as LHS divided by RHS.
The Bnet metric is calculated by first plotting a kernel density estimate of the BDamage values of Glu and Asp side chain oxygen atoms. The area under the curve to the left- (LHS) and right-hand side (RHS) of the median BDamage value of all atoms in the structure is then measured, and Bnet is calculated as LHS divided by RHS.
RABDAM will calculate the BDamage, Bnet and Bnet-percentile metrics for any standard format PDB or mmCIF file to identify potential individual sites, plus the total extent, of specific radiation damage within the structure. BDamage values can be calculated for any macromolecule type; because the Bnet and Bnet-percentile metrics are based upon a damage artefact characterised in cryo-temperature protein crystal structures, accordingly they should only be calculated for such structures.
-
RABDAM can be downloaded/cloned from GitHub. You can then either run RABDAM as a script from the RABDAM directory, or alternatively you can install RABDAM as a package (which can be run from any directory) by navigating to the RABDAM directory and executing:
python setup.py install -
RABDAM is incorporated as part of the CCP4 software suite. It is currently available as a command line package.
RABDAM requires Python 3.8 or above (owing to its dependency on cctbx). In addition, it is dependent upon the following packages:
- cctbx
- numpy >= 1.15.0
- matplotlib >= 2.2.0
- scipy >= 1.1.0
- pandas >= 0.24.1
To check whether your computer is missing any of the packages required to run RABDAM, execute:
python rabdam.py --dependencies
RABDAM will take approximately 20 sec to run a 100 kDa structure on a single processor (as estimated from tests performed under a Macintosh Operating System on an Apple M2 processor). It is compatible with Windows, Macintosh and Linux operating systems.
RABDAM can be run on any standard format PDB or mmCIF file of a single model of your crystal structure of interest (specifically, it requires the ATOM/HETATM records, plus the records specifying the space group, resolution, and temperature). Note that because BDamage is a per-atom metric, it should only be calculated for structures for which B-factor values have been refined per-atom, and close to/after you've finished refinement. Furthermore, owing to the correlation between B-factor and occupancy values, the only non-ligand atoms subject to occupancy refinement should be those in alternate conformers (whose occupancy should sum to 1).
RABDAM can be run either as a script or as a package (see the Installation section for further details). The example commands provided below are for running the program as a script. If you are running RABDAM as a package, simply replace python rabdam.py with rabdam.
RABDAM is a command line program. There are three main command line flags that control the program run:
-i/--input-f/--pdb_file-r/--run
The -i and -f flags control the input to the program. One of these two mutually exclusive flags is required for RABDAM to run.
The -i flag is used to specify the path to an input txt file that lists your selected program parameter values (see the "Constructing an input file" section below for details of what this input file should include).
python rabdam.py -i /path/to/input_file.txt
Alternatively, if you wish to perform a run of RABDAM using entirely default parameter values, it is possible to run RABDAM without an input file; in this case the -f flag is used to provide RABDAM with either a 4 character PDB accession code (XXXX), or a file path (/path/to/pdb_file.pdb or /path/to/mmcif_file.cif), of the model to be analysed:
python rabdam.py -f XXXX
python rabdam.py -f /path/to/pdb_file.pdb
python rabdam.py -f /path/to/mmcif_file.cif
It is possible to specify multiple inputs following the -f flag, e.g.:
python rabdam.py –f /path/to/pdb_file_1.pdb XXXX /path/to/mmcif_file_2.cif YYYY ZZZZ
Importantly, note that file path(s) must not contain any spaces.
-r is an optional flag that controls the output from the program.
The -r flag can be used to instruct RABDAM to run to completion (default), or to stop/start part way through its full run. RABDAM is structured such that it writes the BDamage values calculated for an input model to a dataframe; this dataframe is then used to write the program output files. Through use of the -r flag it is possible to instruct RABDAM to stop (-r df / -r dataframe) or start (-r analysis) its run following dataframe construction. This option will save time if for example you wish to change which output files are generated (which can be controlled using parameters specified in the input txt file - see the “Constructing an input file” section below) without changing the BDamage distribution itself.
In addition, there are two supplementary command line flags:
--dependencies--version
The --dependencies flag directs the program to test whether the system it is being run on has the necessary Python packages installed for RABDAM to run to completion. The --version flag prints the version of RABDAM you are running.
If you wish to run RABDAM with non-default parameter values, you will need to provide the program with an input file specifying your selected parameter values:
- The name of the PDB/mmCIF file(s) to be analysed
Either a 4 character PDB accession code, or a file path (which may not include spaces). It is possible to run multiple structures from a single input file by listing the names of each of those structures separated by commas (see below). This is the only parameter not stipulated by a keyword, and which does not have a default value.
- The output directory, outputDir
The directory in which you would like the program output files to be written. If not specified, this defaults to the current working directory.
- Option to ignore recognised errors encountered during the program run, batchContinue
When set to True, if RABDAM encounters a recognised program error during a run, it either skips to the next model or continues with the run (depending on whether the error raised is "exit" or "pause"). When set to False (default), the program terminates/pauses with exit/pause errors, respectively.
- Option to overwrite pre-existing files with the same name as the new output files, overwrite
Directs the program, if it encounters files of the same name as the output files it is going to write already present in the output directory, to always overwrite these pre-existing files (True), or to pause and require user input to decide whether to overwrite or not (False, default behaviour).
- Option to specify the output files generated, outfiles
When set to "all" (default), directs the program to write all output files. When set to "bnet", directs the program to just record Bnet and Bnet-percentile values.
- Option to check the input model passes the recommended filters for Bnet and Bnet-percentile calculation, filter
When set to True (default value is False), directs RABDAM to check the input model meets the recommended filters for Bnet and Bnet-percentile calculation. These filters are: - Rfree < 0.4 - Resolution <= 3.5 - 80 K <= temperature <= 120 K - No Glu or Asp residues whose occupancy across all conformers is less than 1 - Contains protein - Has >= 20 Glu/Asp side chain oxygen atoms - Refined with per-atom B-factors
- Option to specify model temperature, temperature
Allows user to specify the temperature the dataset was collected at. Default is None, i.e. RABDAM will search the input model file for the temperature.
- Option to specify model resolution, resolution
Allows user to specify the model's resolution (in Angstroms). Default is None, i.e. RABDAM will search the input model file for the resolution.
Note that if a parameter is not specified in the input file, it will take its default value in the RABDAM run.
Below is an example input file instructing RABDAM to analyse the GH7 cellobiohydrolase structures 5MCC and 5MCD, writing the output files to the directory C:\Users\UserName\Documents\RABDAM_test_output, and checking these structures pass the recommended filters for Bnet and Bnet-percentile calculation. All other parameters are set to their default values.
5MCC, 5MCD,
outputdir=C:\Users\UserName\Documents\RABDAM_test_output,
batchContinue=False,
overwrite=False,
outfiles=all,
filter=True,
temperature=None,
resolution=None
Please email kathryn.l.shelley@gmail.com.
- Kathryn Shelley
- Tom Dixon
- Jonny Brooks-Bartlett
This software was developed in the lab of Professor Elspeth Garman at the University of Oxford.
The RABDAM software is described in:
- Shelley KL, Dixon TPE, Brooks-Bartlett JC & Garman EF (2018) RABDAM: quantifying specific radiation damage in individual protein crystal structures. J Appl Cryst 51: 552-559 https://doi.org/10.1107/S1600576718002509
The BDamage metric is defined and validated in:
- Gerstel M, Deane CM & Garman EF (2015) Identifying and quantifying radiation damage at the atomic level. J Synchrotron Radiat 22: 201-212. https://doi.org/10.1107/S1600577515002131
The Bnet and Bnet-percentile metrics are defined and validated in:
- Shelley KL & Garman EF (2022) Quantifying and comparing radiation damage in the Protein Data Bank. Nat Commun 13: 1314. https://doi.org/10.1038/s41467-022-28934-0
RABDAM is distributed within the CCP4 software suite:
- Agirre J, Atanasova M, Bagdonas H, Ballard CB, Baslé A, Beilsten-Edmands J, Borges RJ, Brown DG, Burgos-Mármol JJ, Berrisford JM, Bond PS, Caballero I, Catapano L, Chojnowski G, Cook AG, Cowtan KD, Croll TI, Debreczeni JE, Devenish NE, Dodson EJ, Drevon TR, Emsley P, Evans G, Evans PR, Fando M, Foadi J, Fuentes-Montero L, Garman EF, Gerstel M, Gildea RJ, Hatti K, Hekkelman ML, Heuser P, Hoh SW, Hough MA, Jenkins HT, Jiménez E, Joosten RP, Keegan RM, Keep N, Krissinel EB, Kolenko P, Kovalevskiy O, Lamzin VS, Lawson DM, Lebedev AA, Leslie AGW, Lohkamp B, Long F, Malý M, McCoy AJ, McNicholas SJ, Medina A, Millán C, Murray JW, Murshudov GN, Nicholls RA, Noble MEM, Oeffner R, Pannu NS, Parkhurst JM, Pearce N, Pereira J, Perrakis A, Powell HR, Read RJ, Rigden DJ, Rochira W, Sammito M, Sánchez Rodríguez F, Sheldrick GM, Shelley KL, Simkovic F, Simpkin AJ, Skubak P, Sobolev E, Steiner RA, Stevenson K, Tews I, Thomas JMH, Thorn A, Triviño Valls J, Uski V, Usón I, Vagin A, Velankar S, Vollmar M, Walden H, Waterman D, Wilson KS, Winn MD, Winter G, Wojdyr M & Yamashita K (2023) TheCCP4 suite: integrative software formacromolecular crystallography. Acta Cryst. D 79: 449-461. https://doi.org/10.1107/S2059798323003595