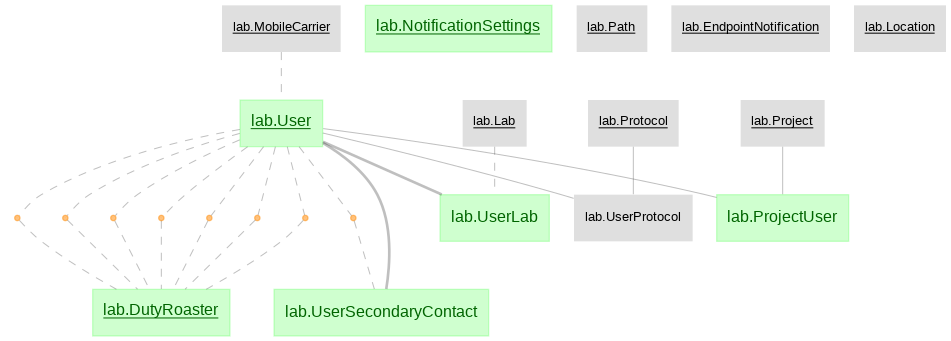
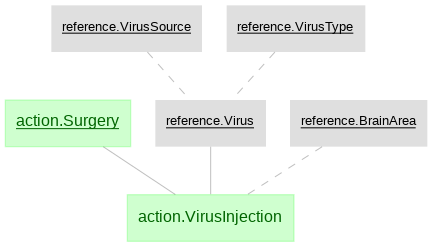
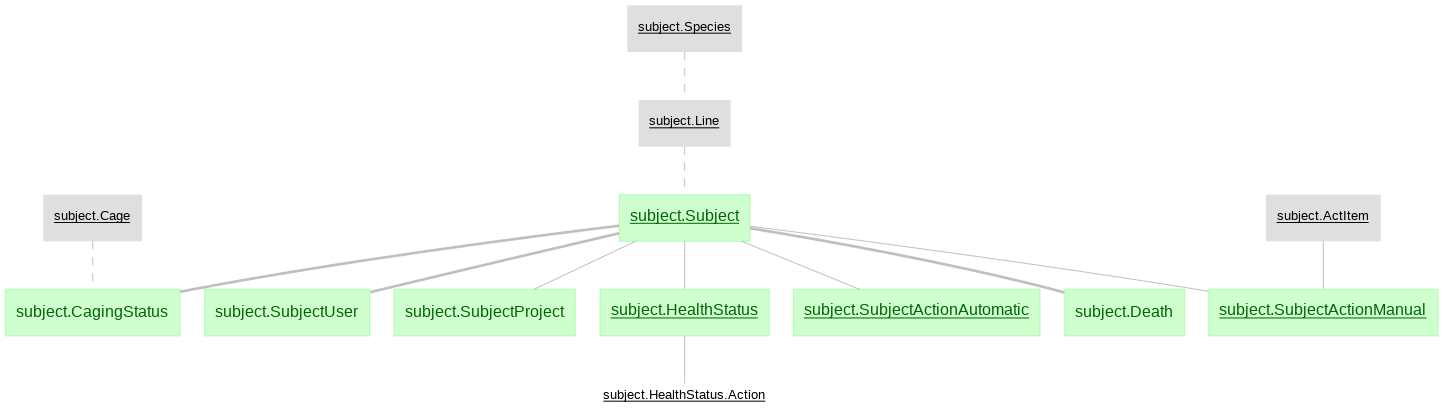
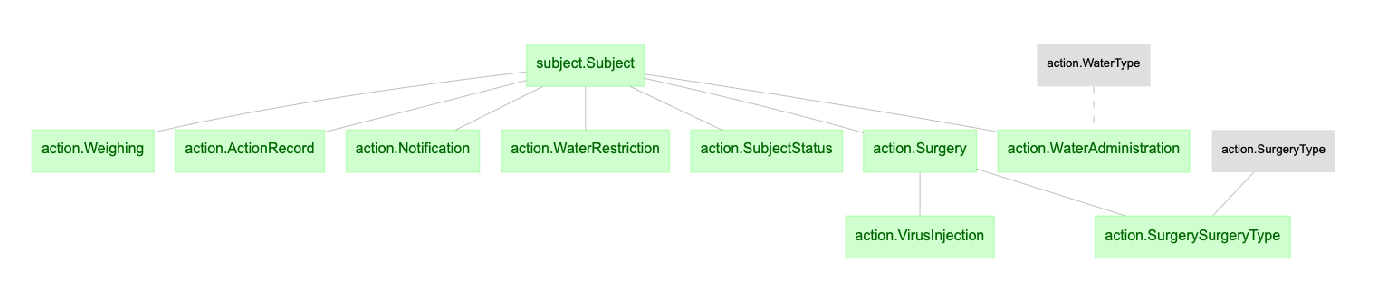
The python data pipeline defined with DataJoint for U19 projects
The data pipeline is mainly ingested and maintained with the matlab repository: https://github.com/BrainCOGS/U19-pipeline-matlab
This repository is the mirrored table definitions for the tables in the matlab pipeline.
- Install conda on your system: https://conda.io/projects/conda/en/latest/user-guide/install/index.html
- If running in Windows get git
- (Optional for ERDs) Install graphviz
- Open a new terminal
- Clone this repository:
git@github.com:BrainCOGS/U19-pipeline_python.git- If you cannot clone repositories with ssh, set keys
- Create a conda environment:
conda create -n u19_datajoint_env python==3.7. - Activate environment:
conda activate u19_datajoint_env. (Activate environment each time you use the project) - Change directory to this repository
cd U19_pipeline_python. - Install all required libraries
pip install -e . - Datajoint Configuration:
jupyter notebook notebooks/00-datajoint-configuration.ipynb
We have created some tutorial notebooks to help you start working with datajoint
- Querying data (Strongly recommended)
jupyter notebook notebooks/tutorials/1-Explore U19 data pipeline with DataJoint.ipynb
- Building analysis pipeline (Recommended only if you are going to create new databases or tables for analysis)
jupyter notebook notebooks/tutorials/2-Analyze data with U19 pipeline and save results.ipynbjupyter notebook notebooks/tutorials/3-Build a simple data pipeline.ipynb
Ephys element and imaging element require root paths for ephys and imaging data. Here are the notebooks showing how to set up the configurations properly.
There are several data files (behavior, imaging & electrophysiology) that are referenced in the database To access thse files you should mount PNI file server volumes on your system. There are three main file servers across PNI where data is stored (braininit, Bezos & u19_dj)
- From Windows Explorer, select "Map Network Drive" and enter:
\\cup.pni.princeton.edu\braininit\ (for braininit)
\\cup.pni.princeton.edu\Bezos-center\ (for Bezos)
\\cup.pni.princeton.edu\u19_dj\ (for u19_dj) - Authenticate with your NetID and PU password (NOT your PNI password, which may be different). When prompted for your username, enter PRINCETON\netid (note that PRINCETON can be upper or lower case) where netid is your PU NetID.
- Select "Go->Connect to Server..." from Finder and enter:
smb://cup.pni.princeton.edu/braininit/ (for braininit)
smb://cup.pni.princeton.edu/Bezos-center/ (for Bezos)
smb://cup.pni.princeton.edu/u19_dj/ (for u19_dj) - Authenticate with your NetID and PU password (NOT your PNI password, which may be different).
- Follow extra steps depicted in this link: https://npcdocs.princeton.edu/index.php/Mounting_the_PNI_file_server_on_your_desktop
Here are some shortcuts to common used data accross PNI
Sue Ann's Towers Task
- Imaging: /Bezos-center/RigData/scope/bay3/sakoay/{protocol_name}/imaging/{subject_nickname}/
- Behavior: /braininit/RigData/scope/bay3/sakoay/{protocol_name}/data/{subject_nickname}/
Lucas Pinto's Widefield
- Imaging /braininit/RigData/VRwidefield/widefield/{subject_nickname}/{session_date}/
- Behavior /braininit/RigData/VRwidefield/behavior/lucas/blocksReboot/data/{subject_nickname}/
Lucas Pinto's Opto inactivacion experiments
- Imaging /braininit/RigData/VRLaser/LaserGalvo1/{subject_nickname}/
- Behavior /braininit/RigData/VRLaser/behav/lucas/blocksReboot/data/{subject_nickname}/
- Mount needed file server
- Connect to the Database
- Create a structure with subject_fullname and session_date from the session
key['subject_fullname'] = 'koay_K65'
key['session_Date'] = '2018-02-05' - Fetch filepath info:
data_dir = (acquisition.SessionStarted & key).fetch('remote_path_behavior_file')
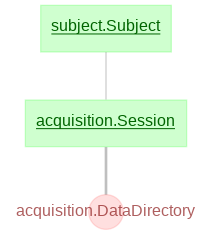
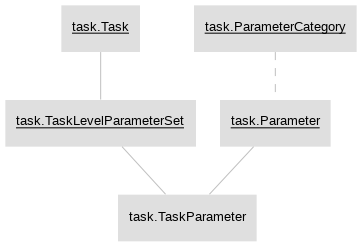
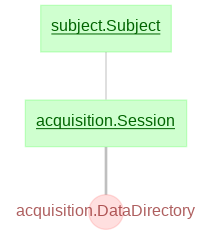
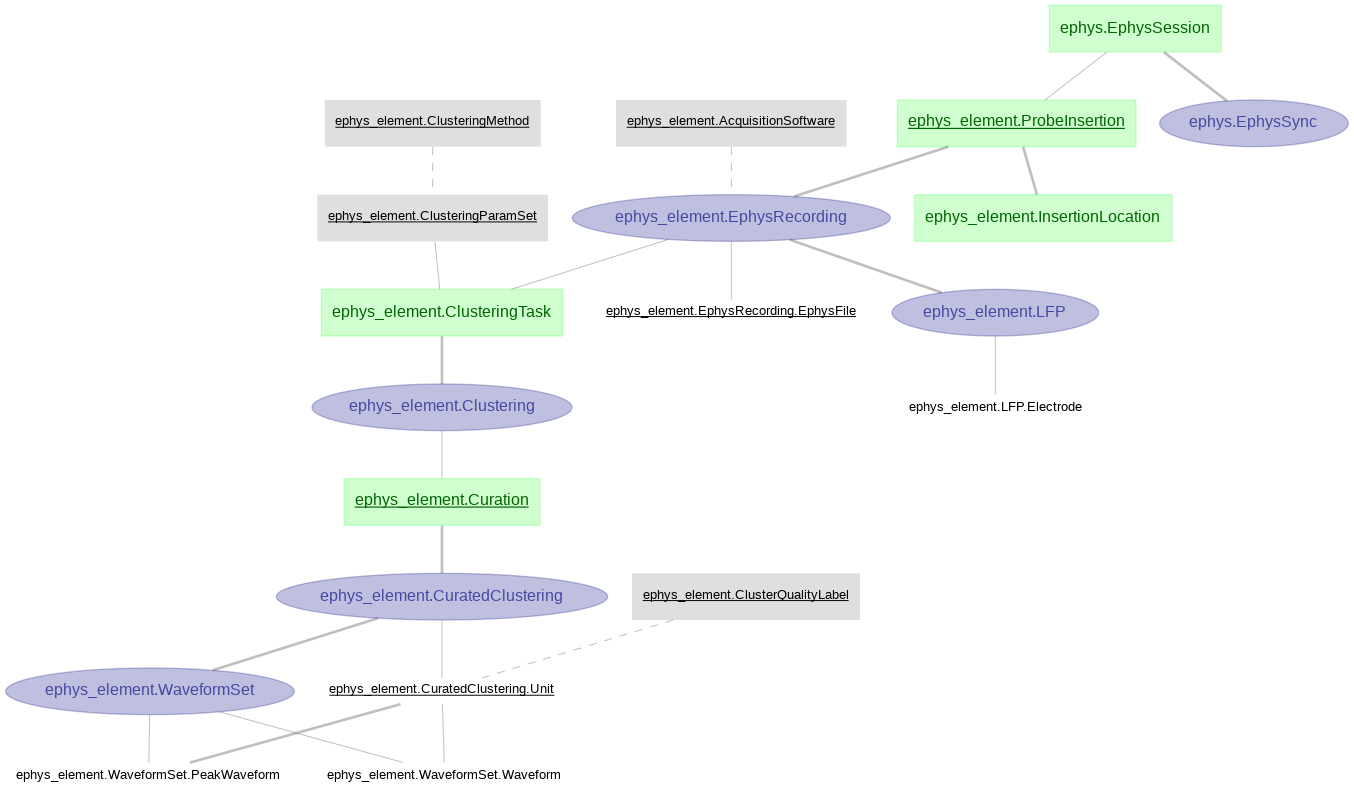
Currently, the main schemas in the data pipeline are as follows:
- lab
- reference
- subject
- action
- acquisition
- task
- behavior
Behavior data for Towers task.
- ephys_element
Ephys related tables were created with DataJoint Element Array Ephys, processing ephys data aquired with SpikeGLX and pre-processed by Kilosort2. For this pipeline we are using the (acute) ephys module from element-array-ephys.
-
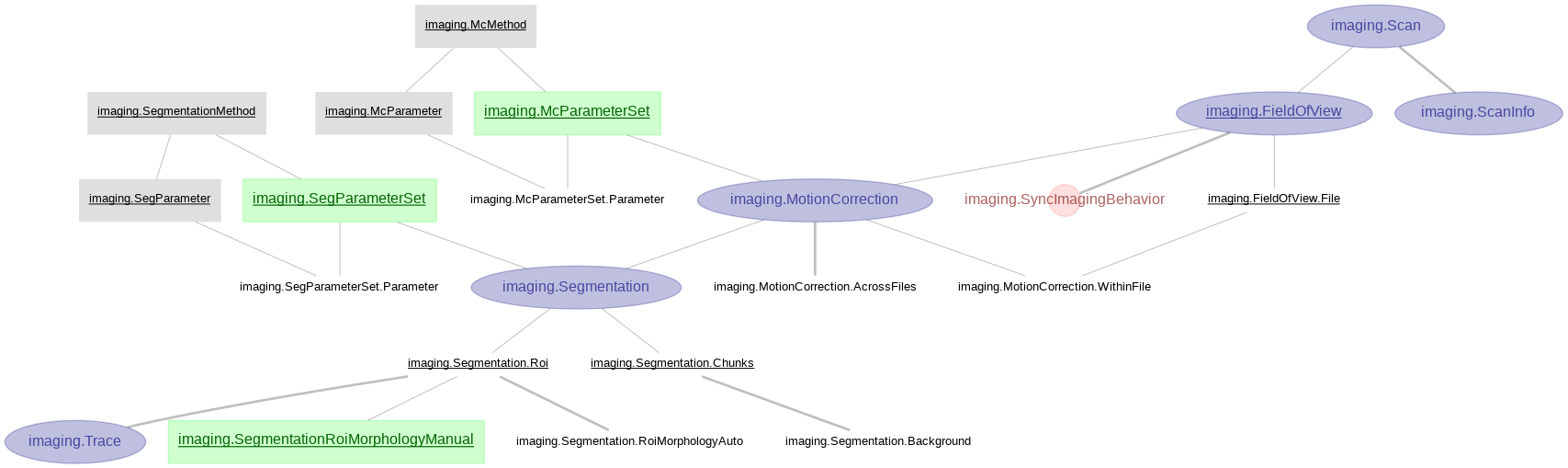
imaging Imaging pipeline processed with customized algorithm for motion correction and CNMF for cell segmentation in matlab.
-
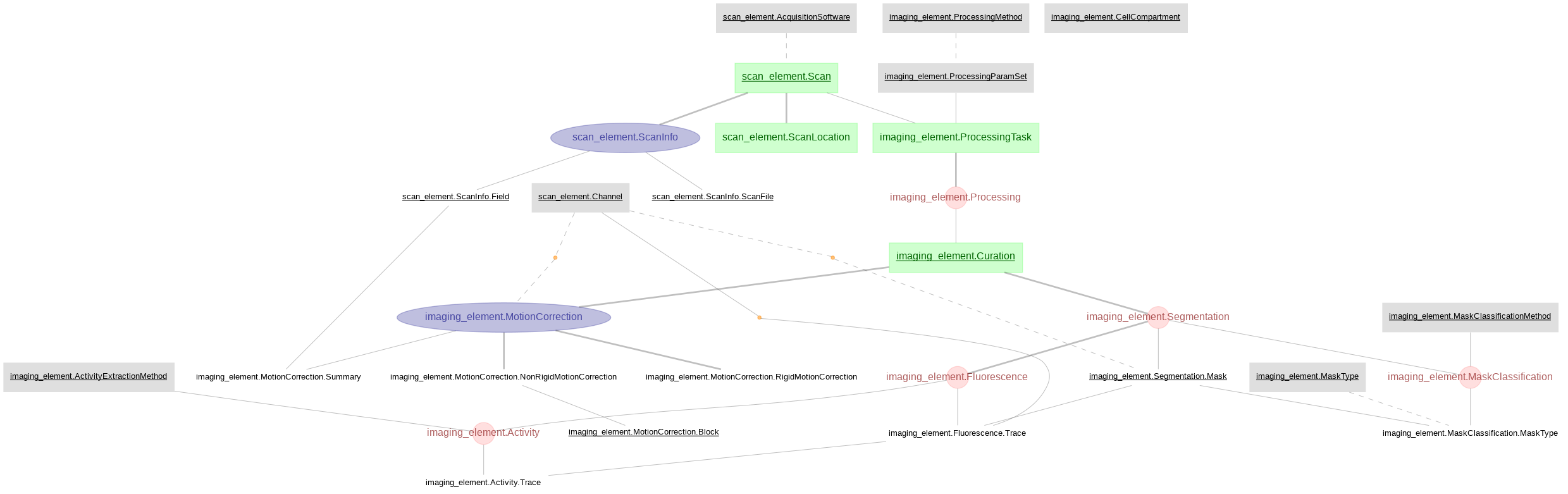
scan_element and imaging_element
Scan and imaging tables created with DataJoint Element Calcium Imaging, processing imaging data acquired with ScanImage and pre-processed by Suite2p.
Import datajoint as follows:
import datajoint as djdj.Table._update(schema.Table & key, 'column_name', 'new_data')
table.heading.attributes.keys()
This also works on a query object:
schema = dj.create_virtual_module("some_schema","some_schema")
query_object = schema.Sample() & 'sample_name ="test"'
query_object.heading.attributes.keys()