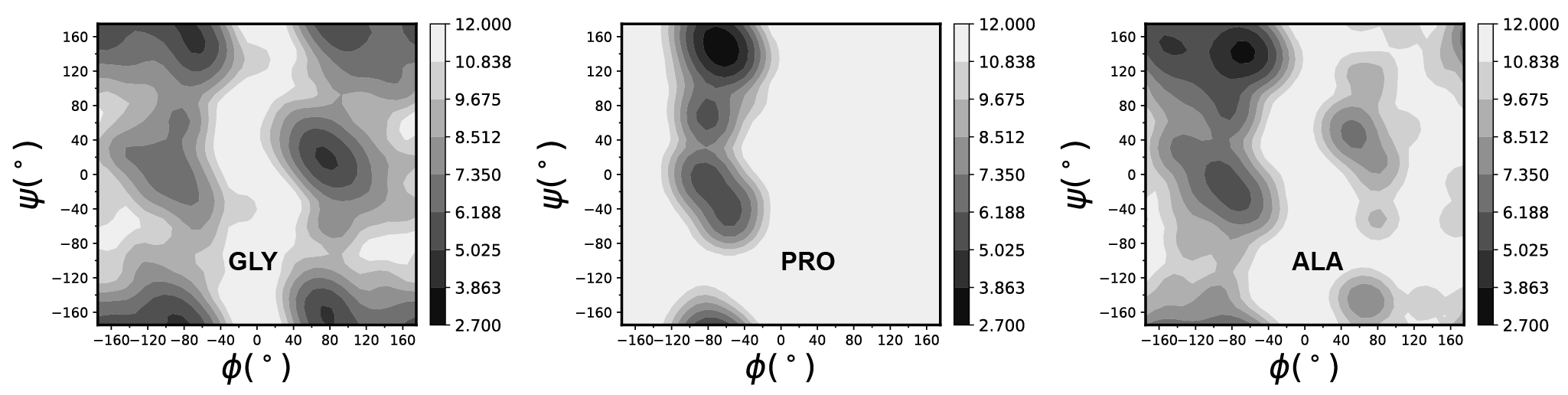
The protein coil library consists of fragments that lie outside regular secondary structures. Secondary structure is formally defined by the pattern of hydrogen bonds between the amino hydrogen and carboxyl oxygen atoms in the peptide backbone. To construct a proper coil library, the following steps have been implemented:
- Firstly, all protein crystal structures were downloaded from the Protein Data Bank, but only those with a resolution of less than 2.0 Å, an observed R factor value of less than 0.2, or a free R factor value of less than 0.2 were used in the next step.
- Secondly, we used BLAST to calculate sequence identity and applied a 50% sequence identity cutoff to filter the proteins.
- Then, we used
DSSPto assign the secondary structure to all residues in the proteins. Any residue assigned to a secondary structure was discarded. Pre-Pro (preceding proline) residues were also excluded to avoid the neighbor residue effect (NRE) of proline. - To validate the protein coil library, we generated Ramachandran plots for GLY, ALA, and PRO. These plots closely resemble those presented in previous studies
$^{[1]}$ .
As mentioned above, only proteins with a resolution less than 2.0 Å, an R value less than 0.2, and generated by diffraction will be adopted. Therefore, one would need to run GetResolutionAndRvalue.py to obtain the associated data. To calculate sequence identity, one should first run ExtractSequencesFromCIF.py to obtain the protein sequences. It may be better to separate all protein into multiple more smaller parts to deal data more quickly.
Create a database for all sequences in Proteins Bank Data
makeblastdb -in totseq.fasta -dbtype prot -title totseqdata -out totseqdata
-in: the file that contains all sequences.-dtype: the type of database: 'prot' refers to the protein type.-title: the name of generated database.
Use BLASTP for protein sequence matching, extract the identity information from the results, and generate the sequence identity matrix
blastp -db data -query query.fasta -outfmt 6 -out blastp.txt
-query: the query sequences.-outfmt: Specify the format of the output data, which includes 12 types. Selecting type 6 outputs the data as a table.-out: The output file. If not specified, the result will be output to the console.
output:
6K9J_A 6K9J_A 100.000 104 0 0 1 104 1 104 2.25e-77 212
6K9J_A 9FE0_D 29.630 27 17 1 42 66 80 106 3.1 15.0
6K9J_A 9FE0_D 32.558 43 16 3 66 101 301 337 5.1 14.2
6K9J_A 9FE0_B 29.630 27 17 1 42 66 80 106 3.1 15.0
6K9J_A 9FE0_B 32.558 43 16 3 66 101 301 337 5.1 14.2
sequence identity is located at third column. It may be better to separate all protein sequences into multiple parts to generate the sequence identity matrix more quickly. Then, you can use the Construct_sequence_database_w_identity.py script to construct a library with a specific sequence identity cutoff in blastb_fulfilseqidentity.
After performing the above steps, one needs to run ReadMMCIF_biophy_exclude_notfulfil_seqiden.py to construct the Protein Coil Library. The Protein Coil Library is archived as proteinCoilLibrary.tar.gz.
a 2D Gaussian kernel estimator was used to extract
Here ramachandran folder
- [1] Jiang, F., Han, W., & Wu, Y. D. (2010). Influence of side chain conformations on local conformational features of amino acids and implication for force field development. The Journal of Physical Chemistry B, 114(17), 5840-5850.
- [2] Jiang, F., Zhou, C. Y., & Wu, Y. D. (2014). Residue-specific force field based on the protein coil library. RSFF1: modification of OPLS-AA/L. The Journal of Physical Chemistry B, 118(25), 6983-6998.