Comparing foundation models and nnU-Net for segmentation of primary brain lymphoma on clinical routine post-contrast T1-weighted MRI
This repository contains material associated to this paper.
It contains:
- link to trained models for segmentation of lymphoma from post-constrast T1-weighted MRI (link)
- link to trained models for segmentation of enhancing tumor in MSD-BraTS datasets (link)
- code and material for reproducing the experiments on MSD-BraTS (link)
If you use this material, we would appreciate if you could cite the following reference.
- Guanghui Fu, Lucia Nichelli, Dario Herran, Romain Valabregue, Agusti Alentorn, Khê Hoang-Xuan, Caroline Houillier, Didier Dormont, Stéphane Lehéricy, Olivier Colliot. Comparing foundation models and nnU-Net for segmentation of primary brain lymphoma on clinical routine post-contrast T1-weighted MRI. Preprint, https://hal.science/hal-04447318.
-
Install SAM repository:
pip install git+https://github.com/facebookresearch/segment-anything.git -
Install UniverSeg repository:
pip install git+https://github.com/JJGO/UniverSeg.git -
Dataset download (from medical segmentation decathlon website): https://drive.google.com/file/d/1A2IU8Sgea1h3fYLpYtFb2v7NYdMjvEhU/view?usp=drive_link
-
Model checkpoints download: Please download SAM and MedSam models from the following links, and put these checkpoints to
sam_experiment/checkpointspath.- SAM, vit-b: https://dl.fbaipublicfiles.com/segment_anything/sam_vit_b_01ec64.pth
- SAM, vit-h: https://dl.fbaipublicfiles.com/segment_anything/sam_vit_h_4b8939.pth
- SAM, vit-l: https://dl.fbaipublicfiles.com/segment_anything/sam_vit_l_0b3195.pth
- MedSAM, vit-b: https://drive.google.com/file/d/1UAmWL88roYR7wKlnApw5Bcuzf2iQgk6_/view?usp=drive_link
To be as similar as possible to lymphoma segmentation task, we selected the T1-weighted MRI after gadolinium injection and focused on the enhancing tumor as the target region. Therefore, we need some basic preprocessing to obtain T1-GD from MRI and select enhancing tumors from the label file of MSD-BraTs dataset.
preprocess/main_preprocess.py:- Split dataset
- Get T1-GD modality from dataset
- Binary label and get enhancing tumor
preprocess/lymphoma_preprocess.py:- Resample to (1,1,1)
- Rescale intensity
- CropOrPad, or Resize to (240, 240, 160)
- Note that, all these operations are performed through TorchIO operations
- Trained model link: https://owncloud.icm-institute.org/index.php/s/2dPGj9hu4Jvk6Qh/download?path=%2F&files=Dataset910_Lymphoma.zip
There is no preprocessing needed for using trained nnU-net model to inference on MSD-BraTS.
- Trained model link: https://owncloud.icm-institute.org/index.php/s/2dPGj9hu4Jvk6Qh/download?path=%2F&files=Dataset912_BRATS.zip
We provide the following contents for reproduction of MSD-BraTS experiments:
- list of subjects of MSD-BraTS that were used (link)
- manual box prompts for SAM and MedSAM models (link)
- support sets for UniverSeg experiments (link)
- code to train nnU-Net models (link)
- code for inference of all models (SAM/MedSAM, UniverSeg, nnU-Net)
- code for computation of metrics and statistical analysis (link)
- Note that, here train means train/validation for nnU-Net.
SAM and MedSAM require prompts. In this paper, we drew box prompts manually.
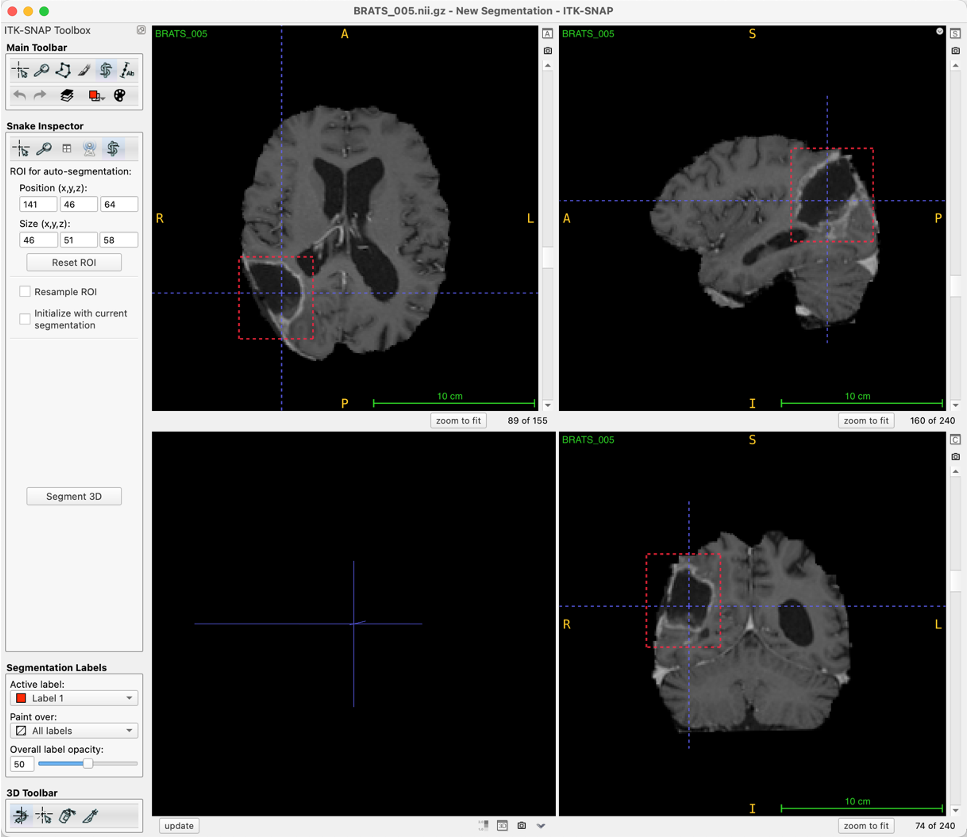
The figure below shows our process of using ITK-SNAP for drawing box prompts.
We also provide screen recording videos during annotation: https://owncloud.icm-institute.org/index.php/s/9LWatZ2xDB9SvE0
In order to reproduce the experiments, you need the coordinates of the box prompts which are given here:
sam_experiment/brats_prompt/3d_manual.csv: The manual annotate box prompt in 3D level
Also we provide the prompts generated from ground-truth.
sam_experiment/brats_prompt/3d_gt.csv: The box prompt generate from ground truth in 3D level.sam_experiment/brats_prompt/2d_gt: The box prompt generate from ground truth in 2D level
The code to generate the boxes from ground truth is provided here:
sam_experiment/label2box_3d.py: The code to generate box prompt from ground truth in 3D level.sam_experiment/label2box_2d.py: The code to generate box prompt from ground truth in 2D slice level.
The different support sets are given here: brats_support_set.zip
The code to build the support sets:
universeg_experiment/universeg_select_support_set.py: this code offers strategies for selecting the support set, a crucial step in preparing data for the UniverSeg [2] model. It includes options to select slices based on their size (largest, smallest, or medium).
Training nnU-Net only requires the following commands. DATASET_ID and FOLD are defined in the same way as nnU-net official codes. See the official code for details.
- Plan and preprocessing:
nnUNetv2_plan_and_preprocess -d DATASET_ID --verify_dataset_integrity- 3D model training:
nnUNetv2_train DATASET_ID 3d_fullres FOLD- 2D model training:
nnUNetv2_train DATASET_ID 2d FOLDsam_experiment/main_sam_manual_prompt.py: it can load the 3D-level manual annotate box prompt, and use SAM/MedSAM to inference.sam_experiment/main_sam_gt_prompt.py: it can load the 3D/2D level box prompt generate from ground truth, and use SAM/MedSAM to inference.
universeg_experiment/main_universeg.py: it contains these three key steps:- Transfer data from 3d to 2D and resize to (128,128)
- Create support set
- Inference the UniverSeg model
Inferencing nnU-Net only requires the following commands. DATASET_ID, INPUT_FOLDER and OUTPUT_FOLDER are defined in the same way as nnU-net official codes. See the official code for details.
- 3D inference:
nnUNetv2_predict -d DATASET_ID -i INPUT_FOLDER -o OUTPUT_FOLDER -f 0 1 2 3 4 -tr nnUNetTrainer -c 3d_fullres -p nnUNetPlans- 2D inference:
nnUNetv2_predict -d DATASET_ID -i INPUT_FOLDER -o OUTPUT_FOLDER -f 0 1 2 3 4 -tr nnUNetTrainer -c 2d -p nnUNetPlansevaluation/eval_bootstrap_ci.py: This code is for evaluation and calculate the 95% bootstrap confidence interval.evaluation/t_test.py: This code is to perform paired T-test.
- SAM [1]: https://github.com/bingogome/samm
- MedSAM [5]: https://github.com/bowang-lab/MedSAM
- UniverSeg [2]: https://github.com/JJGO/UniverSeg
- nnU-Net [4]: https://github.com/MIC-DKFZ/nnUNet
- TorchIO [6]: https://torchio.readthedocs.io/index.html
- Alexander Kirillov, Eric Mintun, Nikhila Ravi, Hanzi Mao, Chloe Rolland, Laura Gustafson, Tete Xiao, Spencer Whitehead, Alexander C Berg, Wan-Yen Lo, et al. Segment anything. In Proc. ICCV 2023, pages 4015–4026, 2023
- Victor Ion Butoi, Jose Javier Gonzalez Ortiz, Tianyu Ma, Mert R Sabuncu, John Guttag, and Adrian V Dalca. UniverSeg: Universal medical image segmentation. In Proc. ICCV 2023, pages 21438–21451, 2023.
- Michela Antonelli, Annika Reinke, Spyridon Bakas, Keyvan Farahani, Annette Kopp-Schneider, Bennett A Landman, Geert Litjens, Bjoern Menze, Olaf Ronneberger, Ronald M Summers, et al. The medical segmentation decathlon. Nature communications, 13(1):4128, 2022.
- Fabian Isensee, Paul F Jaeger, Simon AA Kohl, Jens Petersen, and Klaus H Maier-Hein. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature methods, 18(2):203–211, 2021.
- Ma, Jun, et al. "Segment anything in medical images." Nature Communications 15.1 (2024): 654.
- Pérez-García, Fernando, Rachel Sparks, and Sébastien Ourselin. "TorchIO: a Python library for efficient loading, preprocessing, augmentation and patch-based sampling of medical images in deep learning." Computer Methods and Programs in Biomedicine 208 (2021): 106236.