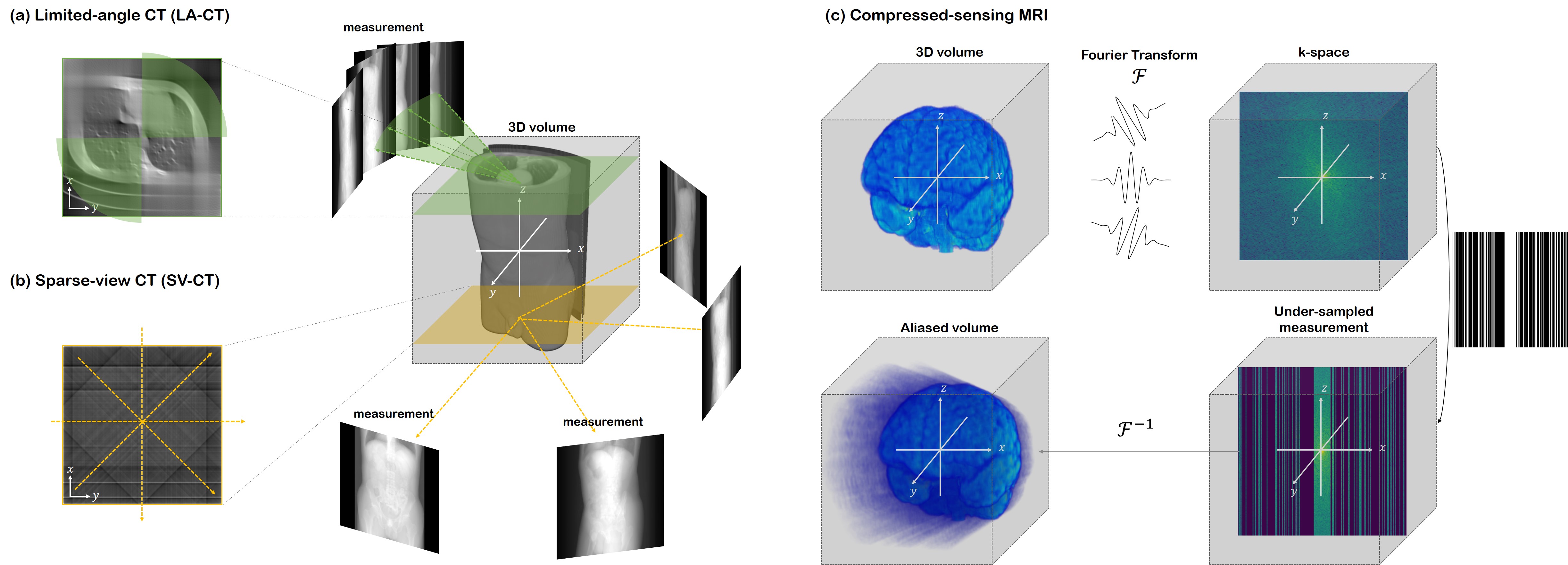
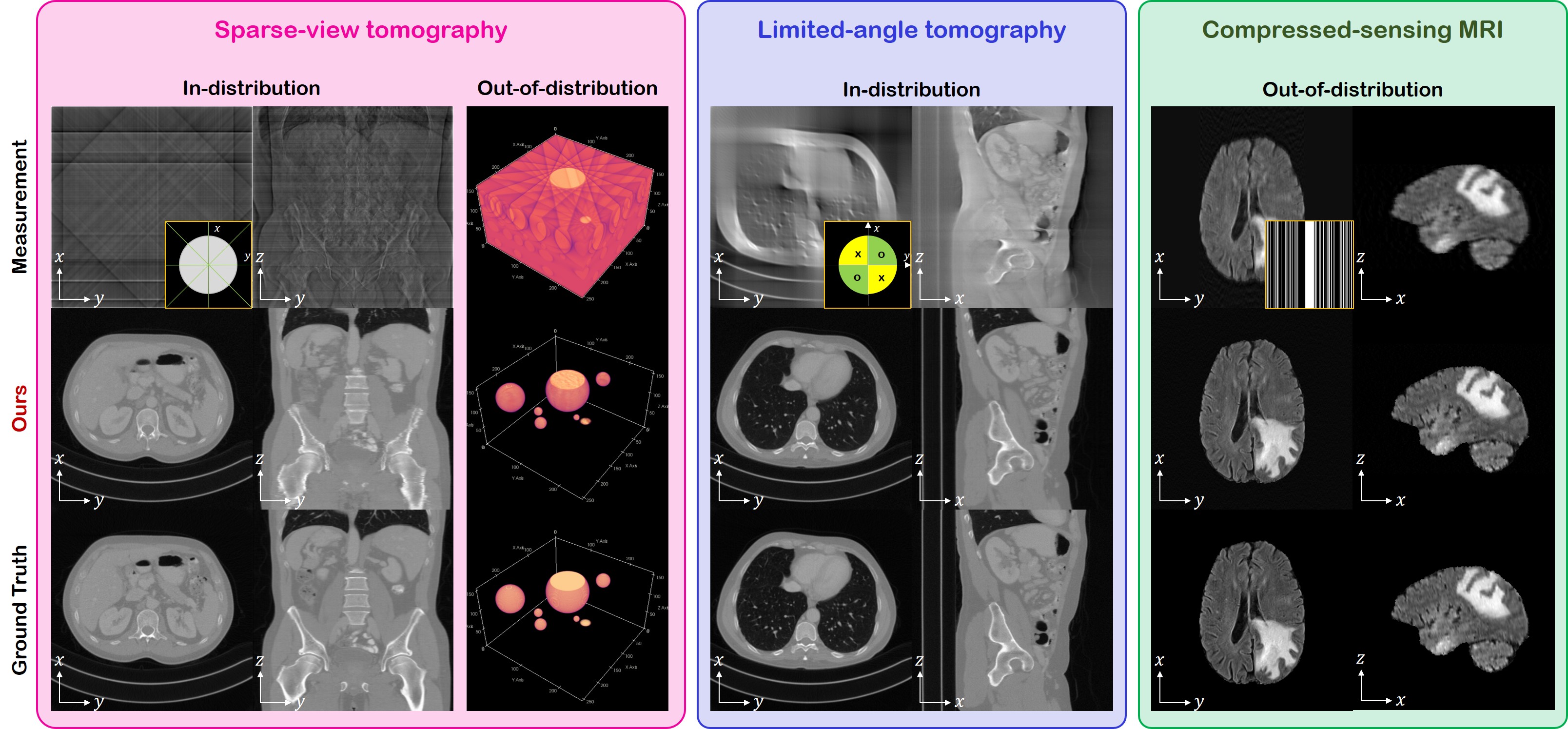
Official PyTorch implementation of DiffusionMBIR, the CVPR 2023 paper "Solving 3D Inverse Problems using Pre-trained 2D Diffusion Models". Code modified from score_sde_pytorch.
- CT experiments
mkdir -p exp/ve/AAPM_256_ncsnpp_continuous
wget -O exp/ve/AAPM_256_ncsnpp_continuous/checkpoint_185.pth https://www.dropbox.com/s/7zevc3eu8xkqx0x/checkpoint_185.pth?dl=1- For MRI experiments
mkdir -p exp/ve/fastmri_knee_320_ncsnpp_continuous
wget -O exp/ve/fastmri_knee_320_ncsnpp_continuous/checkpoint_95.pth https://www.dropbox.com/s/27gtxkmh2dlkho9/checkpoint_95.pth?dl=1(If your system does not have wget installed, you may replace wget -O with curl -L -o.)
- CT experiments (in-distribution)
DATA_DIR=./data/CT/ind/256_sorted
mkdir -p "$DATA_DIR"
wget -O "$DATA_DIR"/256_sorted.zip https://www.dropbox.com/sh/ibjpgo5seksjera/AADlhYqCWq5C4K0uWSrCL_JUa?dl=1
unzip -d "$DATA_DIR"/ "$DATA_DIR"/256_sorted.zip- CT experiments (out-of-distribution)
DATA_DIR=./data/CT/ood/256_sorted
mkdir -p "$DATA_DIR"
wget -O "$DATA_DIR"/slice.zip https://www.dropbox.com/s/h3drrlx0pvutyoi/slice.zip?dl=0
unzip -d "$DATA_DIR"/ "$DATA_DIR"/slice.zip- MRI experiments (out-of-distribution)
DATA_DIR=./data/MRI/BRATS
mkdir -p "$DATA_DIR"
wget -O "$DATA_DIR"/Brats18_CBICA_AAM_1.zip https://www.dropbox.com/s/1a73t58asbqs1mi/Brats18_CBICA_AAM_1.zip?dl=0
unzip -d "$DATA_DIR"/ "$DATA_DIR"/Brats18_CBICA_AAM_1.zip- Make a conda environment and install dependencies
conda env create --file environment.ymlOnce you have the pre-trained weights and the test data set up properly, you may run the following scripts. Modify the parameters in the python scripts directly to change experimental settings.
conda activate diffusion-mbir
python inverse_problem_solver_AAPM_3d_total.py
python inverse_problem_solver_BRATS_MRI_3d_total.pyYou may train the diffusion model with your own data by using e.g.
bash train_AAPM256.shYou can modify the training config with the --config flag.
If you find our work interesting, please consider citing
@InProceedings{chung2023solving,
title={Solving 3D Inverse Problems using Pre-trained 2D Diffusion Models},
author={Chung, Hyungjin and Ryu, Dohoon and McCann, Michael T and Klasky, Marc L and Ye, Jong Chul},
journal={IEEE/CVF Conference on Computer Vision and Pattern Recognition},
year={2023}
}