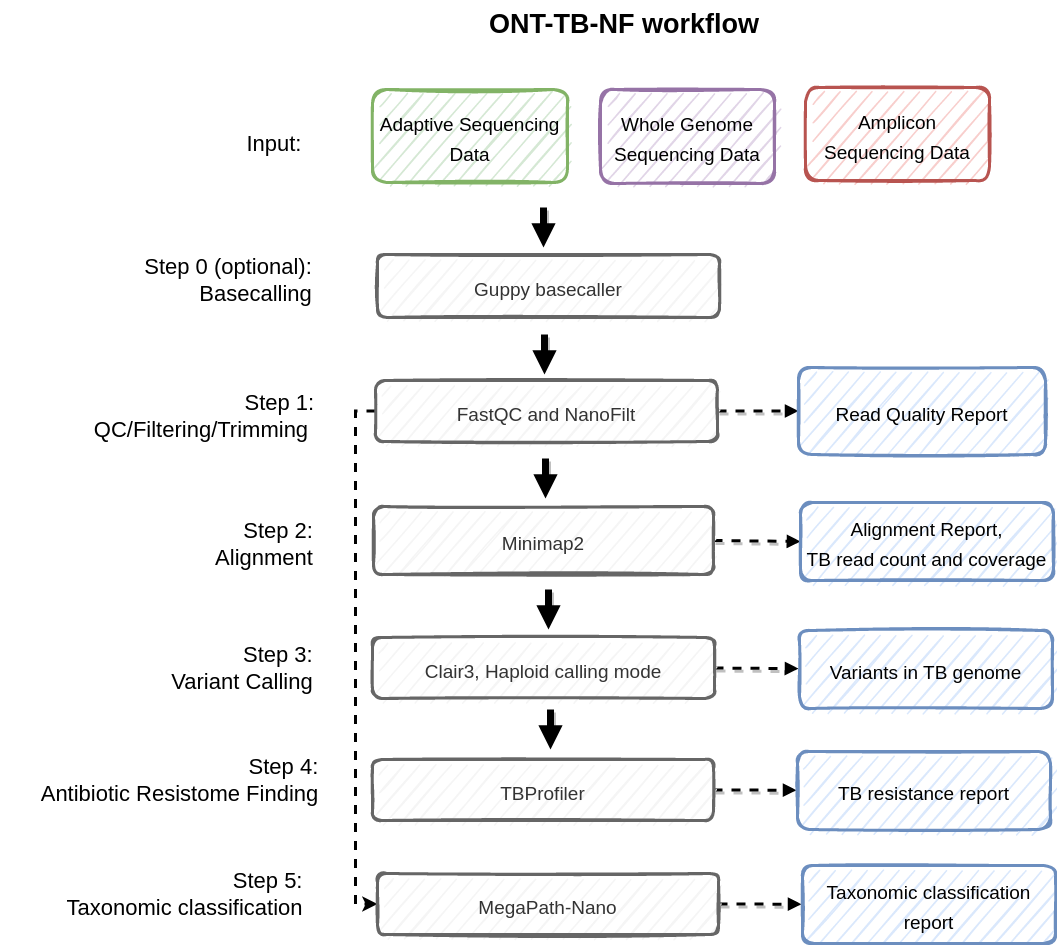
The ONT-TB-NF pipeline is built to detect Mycobacterium tuberculosis (TB) antibiotic-resistance genes from ONT sequencing data.
The input sequencing data can be obtained from various settings of ONT sequencing, including
- Adaptive sequencing (like from readfish or UNCALLED),
- Amplicon sequencing (by amplifying specific regions in the TB genome),
- Standard whole genome sequencing (WGS).
The ONT-TB-NF pipeline includes steps of basecalling, quality control, target regions alignment, variant calling, and antimicrobial resistance prediction.
- One command pipeline from sequencing data to TB analysis report.
- Tailor-made for Adaptive sequencing data, Amplicon, and WGS data.
- Basecalling with ONT's Guppy, the whole pipeline can start from fast5 files.
Install Nextflow by using the following command:
curl -s https://get.nextflow.io | bash
Install required packages with conda and docker:
docker pull hkubal/clair3:v0.1-r12
docker pull quay.io/biocontainers/tb-profiler:4.3.0--pypyh5e36f6f_0
conda create -n ont_tb samtools=1.15.1 minimap2=2.24 nanoplot=1.40.2 mosdepth=0.3.3 flye=2.9.1 nanofilt fastqc bedtools -c bioconda
conda activate ont_tb
# clone ONT-TB-NF
git clone https://github.com/HKU-BAL/ONT-TB-NF.git
cd ONT-TB-NF
Launch the pipeline execution with the following command:
conda activate ont_tb
nextflow run_tb.nf --help
nextflow run_tb_amplicon.nf --help
Make sure you are in the ont_tb environment with the command of conda activate ont_tb.
TB_NF_DIR={ONT-TB-NF PATH}
NF_S=${TB_NF_DIR}/run_tb.nf
SAMPLE_ID={NAME}
FQ={YOUR FQ FILE}
THREADS={THREAD} # threads number, e.g. 16
OUT_DIR={ABSOLUTE OUTPUT PATH} # output path, abolute path required
nextflow run ${NF_S} \
--read_fq ${FQ} \
--sample_name ${SAMPLE_ID} \
--threads ${THREADS} \
--output_dir ${OUT_DIR}
Make sure you are in the ont_tb environment with the command of conda activate ont_tb.
For Amplicon sequencing data, the pipeline needs to be provided with the amplicon bed regions from --amplicon_bed option.
TB_NF_DIR={ONT-TB-NF PATH}
NF_S=${TB_NF_DIR}/run_tb_amplicon.nf
AMPLICON_BED={AMPLICON BED} # your amplicon region
FQ={YOUR FQ FILE}
SAMPLE_ID={NAME}
THREADS={THREAD} # threads number, e.g. 16
OUT_DIR={ABSOLUTE OUTPUT PATH} # output path, abolute path required
nextflow run ${NF_S} \
--read_fq ${FQ} \
--sample_name ${SAMPLE_ID} \
--amplicon_bed ${GENE_BED} \
--threads ${THREADS} \
--output_dir ${OUT_DIR}
FAST5_DIR={Input FAST5 folders}
GUPPY_BASECALLER_PATH={Guppy basecaller path} # e.g. guppy_basecaller
GUPPY_CONFIG={Guppy config file path} # e.g. dna_r10.4_e8.1_sup.cfg
SAMPLE_ID={NAME}
TB_NF_DIR={ONT-TB-NF PATH}
NF_S=${TB_NF_DIR}/run_tb_amplicon.nf # e.g. run_tb.nf or run_tb_amplicon.nf
THREADS={THREAD} # threads number, e.g. 16
OUT_DIR={ABSOLUTE OUTPUT PATH} # output path, abolute path required
nextflow run ${NF_S} --fast5_dir ${FAST5_DIR} --guppy_basecaller_path ${GUPPY_BASECALLER_PATH} --guppy_config_path ${GUPPY_CONFIG} --guppy_options "--device 'cuda:0'" --sample_name ${SAMPLE_ID} --threads ${THREADS} --output_dir ${OUT_DIR}
For apply at WGS and adaptive sequencing, please use the default run_tb.nf pipeline.
For analysis of the Amplicon sequencing data, please use the run_tb_amplicon.nf pipeline.
In general, the ONT-TB-NF pipeline performs the following tasks:
- Basecalling (Guppy, guppy_basecaller)
- Sequencing quality control (FastQC)
- Filtering and Trimming of read (NanoFilt)
- Taxonomic classification (MegaPath-Nano)
- Alignment (minimap2)
- Variant calling (Clair3)
- Antibiotic resistome finding (TBProfiler)
Here is a brief description of output files created for each sample, optional module are labeled with [O]:
[O] Basecalling results at: {YOUR OUTPUR DIR}/0_bc
QC results at: {YOUR OUTPUR DIR}/1_qc
Aligment results at: {YOUR OUTPUR DIR}/2_aln
Variant calling results at: {YOUR OUTPUR DIR}/3_vc
TB analysis report at: {YOUR OUTPUR DIR}/4_tb
[O] taxonomic classification at: {YOUR OUTPUR DIR}/5_mpn