HERGen: Elevating Radiology Report Generation with Longitudinal Data, ECCV 2024.
Fuying Wang, Shenghui Du, and Lequan Yu
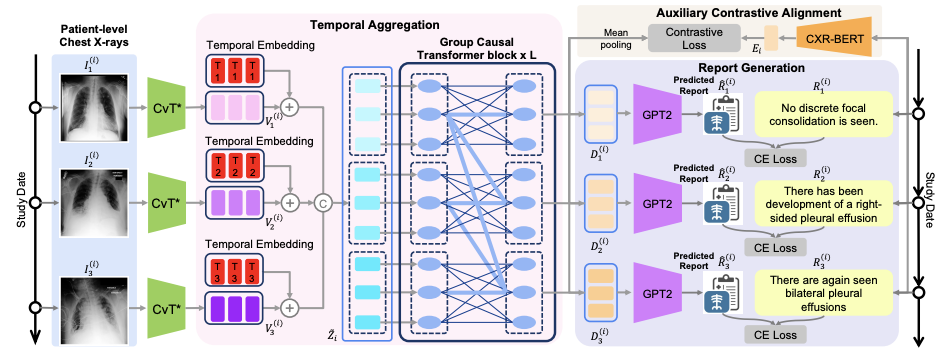
Abstract: Radiology reports provide detailed descriptions of medical imaging integrated with patients’ medical histories, while report writing is traditionally labor-intensive, increasing radiologists’ workload and the risk of diagnostic errors. Recent efforts in automating this process seek to mitigate these issues by enhancing accuracy and clinical efficiency. However, existing automated approaches are based on a single timestamp and often neglect the critical temporal aspect of patients’ imaging histories, which is essential for accurate longitudinal analysis. To address this gap, we propose a novel History Enhanced Radiology Report Generation (HERGen) framework that employs a group causal transformer to efficiently integrate longitudinal data across patient visits. Our approach not only allows for comprehensive analysis of varied historical data but also improves the quality of generated reports through an auxiliary contrastive objective that aligns image sequences with their corresponding reports. More importantly, we introduce a curriculum learning-based strategy to adeptly handle the inherent complexity of longitudinal radiology data and thus stabilize the optimization of our framework. The extensive evaluations across three datasets demonstrate that our framework surpasses existing methods in generating accurate radiology reports and effectively predicting disease progression from medical images.
- 20/11/2024: The first version of HERGen code base is now alive.
Once you clone the repo, please run the following command to create HERGen conda environment.
conda create hergen python=3.10
conda activate hergen
pip install -r requirements.txt
# install our local package: hergen
pip install -e .
We are mainly using the MIMIC-CXR v2.0.0 dataset in our experiments. Please find download the original dataset in the PhysioNet website.
There are 3 necessary steps to prepare the dataset:
-
Please use the MIMIC-CXR preprocessing code in our prior work MGCA to extract the
master.csvfile. -
Download the
mimic_annotation.jsonfrom DCL. The annotation files are in this link. -
Using
hergen/preprocessing/create_temporal_dataset.ipynbto create the our preoprocessed annotations. We are doing two preprocessing:
- Only keep frontal images
- Remove duplicated images in the same study
It results in two annotations:
mimic_annotation.json and longitudinal_mimic_annotation.json for original MIMIC-CXR and longitudinal MIMIC-CXR respectively.
The preprocessed annotation files can also be downloaded in this link.
We are using curriculum learning to conduct training in three stages:
(please specify annotation_file and dataset_dir of your path)
stage 1: Training using individual image-text pairs
CUDA_VISIBLE_DEVICES=0,1 python hergen/tools/train_report_generation.py --num_devices 2 --model_name cvt2distilgpt2 --batch_size 16 \
--annotation_file ANNOTATION_FILE --dataset_dir DATASET_DIR
stage 2: Training the model using additional contrastive alignment
CUDA_VISIBLE_DEVICES=0,1 python hergen/tools/train_report_generation.py --model_name clgen --batch_size 16 --num_devices 2 \
--ckpt_path CKPT_PATH --annotation_file ANNOTATION_FILE --dataset_dir DATASET_DIR
Please change the ckpt_path to the model path in stage 1.
stage 3: Training the whole framework with longitudinal information
CUDA_VISIBLE_DEVICES=0,1 python hergen/tools/train_report_generation.py --model_name temporal_decoder --batch_size 4 --num_devices 2 \
--ckpt_path CKPT_PATH --annotation_file ANNOTATION_FILE --dataset_dir DATASET_DIR
Please change the ckpt_path to the model path in stage 2.
If you find our work useful in your research or if you use parts of our code, please cite our paper:
@article{wang2024hergen,
title={HERGen: Elevating Radiology Report Generation with Longitudinal Data},
author={Wang, Fuying and Du, Shenghui and Yu, Lequan},
journal={arXiv preprint arXiv:2407.15158},
year={2024}
}
The code for HERGen was adapted and inspired by the fantastic works of R2Gen , DCL and cvt2distilgpt2.