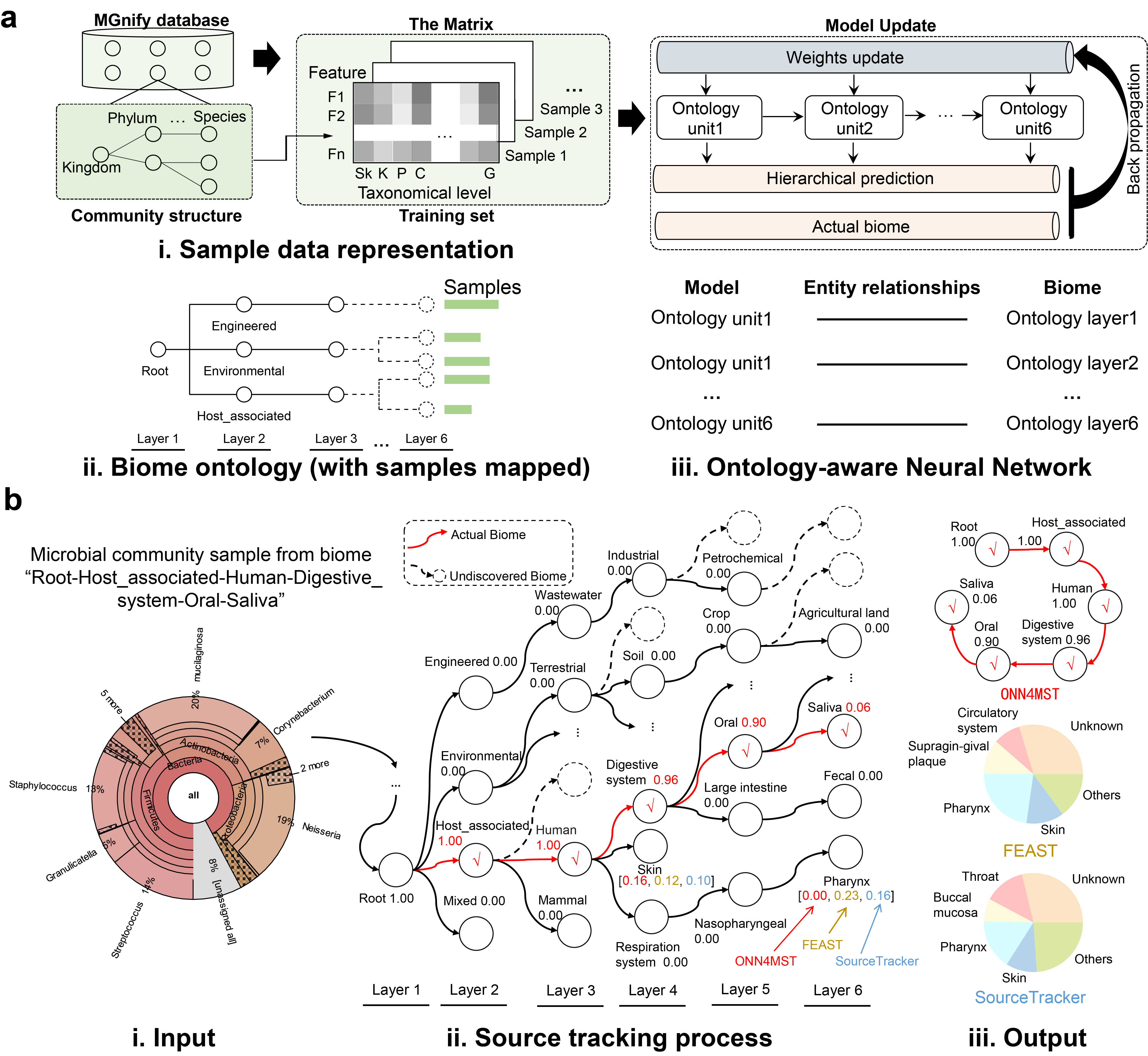
Ontology-aware Neural Network for Microbial Source Tracking!
This program is designed to perform fast and accurate biome source tracking among millions of samples from hundreds of niches. The biome ontology is organized as a tree-like structure, which have six layers. The Neural Network is also organized in six layers, which could produce a hierarchical classification result. Each input sample is represented by a species realtive abundance ".tsv" file, which can be produced by Qiime or obtained from EBI. The output is a biome source ".txt" file, which shows you which biome (niche) the input sample is most likely comes from.
The preprocessing program can make the data preprocess and sample statistical analysis of the Ontology-aware Neural Network easier. For very time-consuming big data calculation, any minor data or program errors can cost days or even weeks. Using this program to check the integrity of all data and error values before processing can greatly reduce the probability of program running errors.
We only provide general model on GitHub. Other models mentioned in our study as well as our training scripts are available upon reasonable request. General models based on all features (44,668 features) and selected features (1462 features) can be found in our releases.
.
├── README.md : Basic information for the repository
├── data : Data files for ONN4MST
│ ├── npzs : ".npz" files and path files for ONN4MST
│ │ ├── GroundWaterSamplesMatrices.npz : ".npz" files for 11 Groundwater samples mentioned in our study
│ │ ├── GroundWaterSamplesPaths.txt : Path to 11 Groundwater samples mentioned in our study
│ │ ├── HumanSamplesMatrices.npz : Path to 10 Human samples mentioned in our study
│ │ └── HumanSamplesPaths.txt : ".npz" files for 10 Human samples mentioned in our study
│ ├── trees : Tree files for ONN4MST
│ │ ├── biome_tree.pkl : Tree file containing serialized biome ontology
│ │ ├── ordered_labels.txt : Biomes involved in the biome ontology
│ │ └── species_tree.pkl : Phylogenetic tree used by ONN4MST
│ └── tsvs : Raw input (".tsv") files for ONN4MST
│ ├── GroundWaterSamplesOTUs.tar.gz : Compressed 11 Groundwater samples
│ └── HumanSamplesOTUs.tar.gz : Compressed 10 Human samples
├── environment.yaml : Dependencies for ONN4MST
├── image : Images used in README
│ ├── Figure1.png
│ ├── Supplementary_Figure5.png
├── src : Source code for ONN4MST
│ ├── document.md : Document for the scripts of ONN4MST
│ ├── dp_utils.py : Utility classes for data preprocessing program
│ ├── gen_ontology.py : Utility functions for the generating of biome ontology
│ ├── graph_builder.py : Utility for computational graph building of ON4MST
│ ├── predicting.py : Utility functions for the prediction function ONN4MST
│ ├── preprocess.py : Main preprocessing program
│ ├── searching.py : Main searching program
│ ├── training.py : Training script of ON4MST
│ ├── testing.py : Testing script of ON4MST
│ └── utils.py : Other utility functions for ONN4MST
└── tmp : Temporal files of ONN4MST
├── 1462FeaturesIndices.npz : ".npz" files containing indices for 1462 selected features
└── error_list : Error list of input data filesFor support using ONN4MST, please email us. Any comments/insights would be greatly appreciated.
-
Unix/Linux operating system
-
At least 2 GB free disk space
-
At least 22 GB RAM
Windows users can easily install ONN4MST through WSL (Windows Subsystem for Linux).
We recommend deploying ONN4MST using git and conda.
# clone this repository
git clone https://github.com/HUST-NingKang-Lab/ONN4MST.git
cd ONN4MST
# download and uncompress our model
wget -c https://github.com/HUST-NingKang-Lab/ONN4MST/releases/download/v0.2/config.tar.gz
tar zxvf config.tar.gz
# configure environment using environment.yaml
conda env create -n ONN4MST -f environment.yaml
conda activate ONN4MSTCheck if the preprocess.py, searching.py and training.py under src/ is executable. If they are not executable, type
chmod +x src/preprocess.py src/searching.py src/training.pyThe input file format of ONN4MST is the ".npz" file. Before ONN, you need to convert the original input ".tsv" file into ".npz" file. The script "src/preprocess.py" could work for it.
If you have successfully converted the ".tsv" file into ".npz" file, then you could run the script "src/searching.py" for biome source tracking. Besides, you need also indicate a trained model. We have provided a well trained model as the default model.
We provide the training script for users to retrain the ONN model with the Combined dataset or self-defined dataset. Before the training, users need to convert the microbial community sample “.tsv” files into ".npz" file via the script "src/preprocess.py". Note that the training and testing datasets need to be generated separately, so users should prepare two dataset files and convert them to ".npz "format (i.e., "trainingdataset.npz" and "testingdataset.npz"). Then, users could run the script "src/training.py" to start the training process.
The model can run either in GPU or CPU mode, we have provided an option -g to indicate that. See Usage for details.
See environment.yaml.
usage: preprocess.py [-h] [-p N_JOBS] [-b BATCH_INDEX] [-s BATCH_SIZE]
[-i INPUT_DIR] [-o OUTPUT_DIR] [-t TREE] [-c COEF]
[--header HEADER]
{check,build,convert,count,merge,select}Basically, the -i, -o and -t arguments for src/proprocess.py are used to specify the input_folder, output_folder and tree_folder.
usage: searching.py [-h] [-g {0,1}] [-gid GPU_CORE_ID] [-s {0,1}] [-t TREE]
[-m MODEL] [-th THRESHOLD] [-mp MAPPING] [-of {1,2,3}]
ifn ofnThe -m and -t arguments for src/searching.py are used to specify model (".json" file, see release page) and biome ontology (".tree" file under config). If you want ONN4MST to run in GPU mode, use -g 1. And the model based on selected features can be accessed by using -m config/model_sf.json with -s 1.
There are several useful arguments (e.g. --batch-Size, --batch and --n_jobs) provided in src/preprocess.py and src/searching.py. You can see them via -h option.
usage: training.py trainingdataset.npz testingdataset.npz model.jsonThere are three arguments for the script training.py, including trainingdataset.npz, testingdataset.npz, and model.json. The arguments of trainingdataset.npz and testingdataset.npz are training and testing dataset files generated by script src/preprocess.py. The argument of model.json is the output model file, which records the architecture and parameters of the ONN model when training completed. We have provided a pre-built model as the default model, it could be found at config/model_df.json, please also see the release page.
Here's a simple guide for using ONN4MST to perform microbial source tracking.
The example data can be found here. Notice that here is a header "# Constructed from biom file" in the first line.
| # Constructed from biom file | ||
|---|---|---|
| # OTU ID | ERR1754760 | taxonomy |
| 207119 | 19.0 | sk__Archaea |
| 118090 | 45.0 | sk__Archaea;k__;p__Thaumarchaeota;c__;o__Nitrosopumilales;f__Nitro... |
| 153156 | 38.0 | sk__Archaea;k__;p__Thaumarchaeota;c__;o__Nitrosopumilales;f__Nitro... |
| 131704 | 1.0 | sk__Archaea;k__;p__Thaumarchaeota;c__Nitrososphaeria;o__Nitrososp... |
| 103181 | 5174.0 | sk__Bacteria |
| 157361 | 9.0 | sk__Bacteria;k__;p__;c__;o__;f__;g__;s__agricultural_soil_bacterium_SC-I-11 |
You need to process your data and organize them into input_folder/biome/tsv_files, just like this.
data/tsvs
└── root-Host_associated-Human
├── ERR1073574_FASTQ_otu.tsv
├── ERR1074236_FASTQ_otu.tsv
└── ERR1077660_FASTQ_otu.tsvGenerally, If you use files from MGnify database, you can just use preprocessing program normally.
src/preprocess.py check -i data/tsvsBut if your ".tsv" files don't have a header in the first line, you need to specify the header argument.
src/preprocess.py check -i data/tsvs --header 0 The file path and error message of all broken data will be saved to error_list and error_msg under tmp folder.
Also, you need to specify the header argument here, take "1" as an example.
src/preprocess.py convert -i data/tsvs -t data/trees -o data/npzs/ --header 1 \
--batch_size 10 --batch_index 0 --n_jobs 1Perform source tracking using ONN4MST
- using model based on all features, in CPU mode.
src/searching.py data/npzs/batch_0.npz searching_result.txt -g 0 -s 0 -t config/microbiome.tree \
-m config/model_df.json -th 0 -of 2 -mp data/npzs/paths.txt- using model based on all features, in GPU mode.
src/searching.py data/npzs/batch_0.npz searching_result.txt -g 1 -s 0 -t config/microbiome.tree \
-m config/model_df.json -th 0 -of 2 -mp data/npzs/paths.txt- using model based on selected features, in CPU mode.
src/searching.py data/npzs/batch_0.npz searching_result.txt -g 0 -s 1 -t config/microbiome.tree \
-m config/model_sf.json -th 0 -of 2 -mp data/npzs/paths.txt- using model based on selected features, in GPU mode.
src/searching.py data/npzs/batch_0.npz searching_result.txt -g 1 -s 1 -t config/microbiome.tree \
-m config/model_sf.json -th 0 -of 2 -mp data/npzs/paths.txtYou need also indicate a mapping file containing paths of your input ".tsv" files using -mp option. Usually, this mapping file is automatically generated by preprocess.py convert. You can find it under the output folder ./data/npzs/.
In the second output format, the predicted sources and their contribution to each sample are given as follows.
>Sample_1 Layer2|root-Engineered,root-Environmental,root-Host_associated...
>Sample_1 Layer2|0.0,4.589534e-06,0.99999374,1.6987236e-06 Layer3|...
>Sample_2 Layer2|root-Engineered,root-Environmental,root-Host_associated...
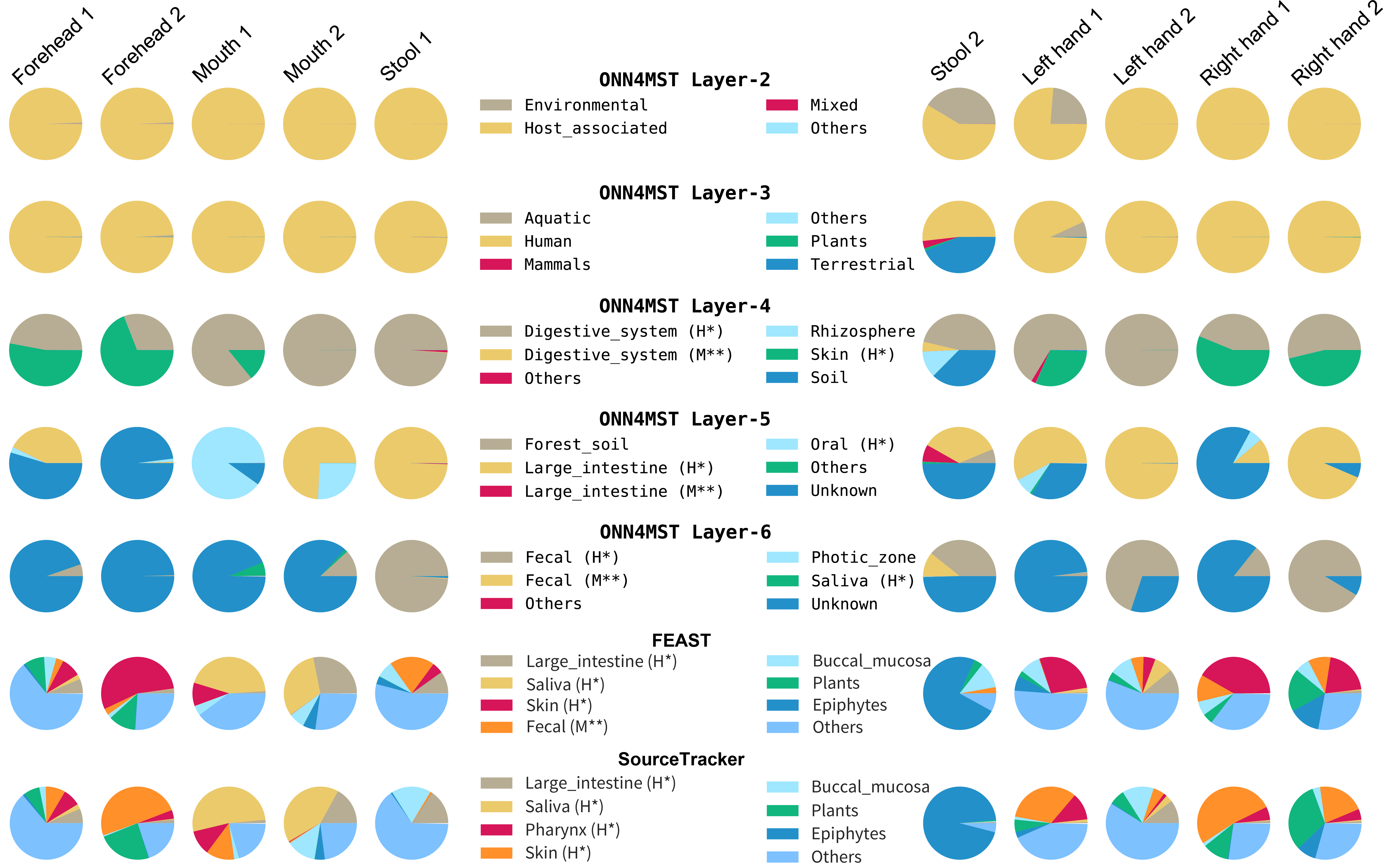
>Sample_2 Layer2|1.5492266e-06,0.00038966027,0.999591,1.7816106e-05 Layer3|...They can be easily visualized into pie charts.
For the evaluation of source tracking methods, please refer to DLMER-Bio.
For easy reproduction, we have converted 11 Groundwater samples into model-acceptable ".npz" files, which can be used to reproduce the experiments "Source tracking of environmental samples from less studied biomes" in our paper. To reproduce the experiment, run the following command after fully configured ONN4MST and its dependencies:
src/searching.py data/npzs/GroundwaterSamplesMatrices.npz ContributionToGroundwaterSamples.txt \
-g 0 -s 0 -t config/microbiome.tree -m config/model_df.json -th 0 -of 3 -mp data/npzs/GroundwaterSamplesPaths.txtTheir MGnify Run ID can be found in GroundwaterSamplesPaths.txt. And their related metadata are given below. The order of samples in output file is in totally agreement with that in GroundwaterSamplesPaths.txt.
| Run ID | Sample description |
|---|---|
| ERR904477 | Well_D |
| ERR904472 | Well_A2 |
| ERR904478 | Well_E |
| ERR904481 | Well_H |
| ERR904475 | Well_C1 |
| ERR904474 | Well_B2 |
| ERR904471 | Well_A1 |
| ERR904480 | Well_G |
| ERR904479 | Well_F |
| ERR904473 | Well_B1 |
| ERR904476 | Well_C2 |
For easy reproduction, we have converted 10 Human samples into model-acceptable ".npz" files, which can be used to reproduce the experiments "Source tracking of samples from closely related human associated biomes" in our paper. To reproduce the experiment, run the following command:
src/searching.py data/npzs/HumanSamplesMatrices.npz ContributionToHumanSamples.txt -g 0 -s 0 \
-t config/microbiome.tree -m config/model_df.json -th 0 -of 3 -mp data/npzs/HumanSamplesPaths.txtTheir MGnify Run ID can be found in HumanSamplesPaths.txt. And their related metadata are given below. The order of samples in output file is in totally agreement with that in HumanSamplesPaths.txt.
| Run ID | Sample description |
|---|---|
| ERR1074236 | American Gut Project Mouth sample (Mouth 2) |
| ERR1074494 | American Gut Project Stool sample (Stool 2) |
| ERR1073574 | American Gut Project Left Hand sample (Left hand 2) |
| ERR1076801 | American Gut Project Left Hand sample (Left hand 1) |
| ERR1074499 | American Gut Project Forehead sample (Forehead 1) |
| ERR1077660 | American Gut Project Right Hand sample (Right hand 2) |
| ERR1074498 | American Gut Project Mouth sample (Mouth 1) |
| ERR1076805 | American Gut Project Right Hand sample (Right hand 1) |
| ERR1074237 | American Gut Project Stool sample (Stool 1) |
| ERR1074238 | American Gut Project Forehead sample (Forehead 2) |
| Name | Organization | |
|---|---|---|
| Hugo Zha | hugozha@hust.deu.cn | Ph.D. Candidate, School of Life Science and Technology, Huazhong University of Science & Technology |
| Hui Chong | huichong.me@gmail.com | Research Assistant, School of Life Science and Technology, Huazhong University of Science & Technology |
| Kang Ning | ningkang@hust.edu.cn | Professor, School of Life Science and Technology, Huazhong University of Science & Technology |