This directory contains the code and resources of the following paper:
"Modeling Gene Regulatory Networks Using Neural Network Architectures" publish in Nature Computational Science (doi:10.1038/s43588-021-00099-8)
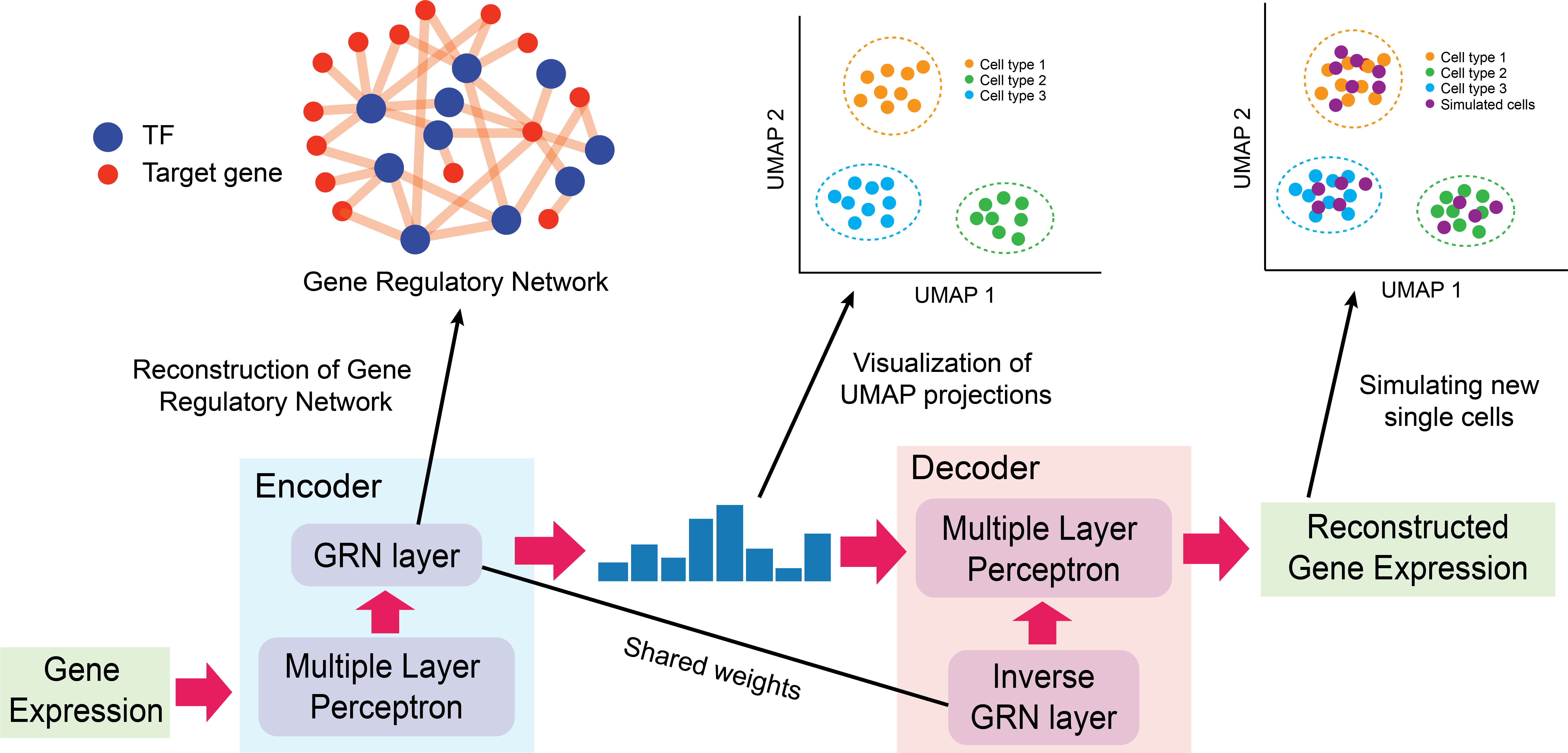
We introduce DeepSEM, a deep-learning-based approach with novel neural network architecture that can infer gene regulatory network, embed scRNA-seq data, and simulate realistic scRNA-seq data by interpreting different modules.
- python 3.7
- pytorch==1.2.0
- scanpy==1.6.0
- numpy==1.14.5
- pandas==1.0.0
- scikit-learn==0.23.2
All dependencies can be installed within a few minutes.
We provide tree tutorial as shown in directory tutorial/{GRN_inference_tutorial.ipynb,Embedding_tutorial.ipynb, Simulation_tutorial.ipynb} for introducing the usage of DeepSEM and reproducing the main result of our paper.
DeepSEM take data as input file in tsv, csv, 10X format, or h5ad format provided by Scanpy (genes in columns and cells in rows for tsv and csv). The output of DeepSEM is varying for different tasks.
- GRN Inference task. A tsv file including TF, Target, and predicted GRN edge importance.
- Embedding. A h5ad file including the embedding genetated by DeepSEM which are shown in "X" of the AnnData and the low dimension representation which are shown in "obsm['X_pca']".
- Simulation. A h5ad file including the simulation result generated by DeepSEM which are shown in "X" of the AnnData.
We also provide default hyper-parameters in main.py. Using -h option or read Hyperparmeter.MD which introduces the hyper-parameters and provides suggestion for hyper-parameter tuning.
Command to run DeepSEM
- Gene Regulation Inference (including cell type specific GRN and cell type non-specific GRN). Note that this is the script for non-ensemble version. We recommend to use ensemble streagy by repeating training process for K times (K=10 in our papaer) and use average of the absolute adjacent matrices as final prediction. Details are shown in tutorial/GRN_inference_tutorial.ipynb.
use --setting test to infer GRN instead of benchmarking.
python main.py --task celltype_GRN --data_file <scGNA-seq path> --save_name <output path> --setting test python main.py --task non_celltype_GRN --data_file <scGNA-seq path> --save_name <output path> --setting test
- Embedding
python main.py --task embedding --data_file <scGNA-seq path> --save_name <output path>
- Simulation
python main.py --task simulation --data_file <scRNA-seq path> --save_name <output path>
- BEELINE https://github.com/Murali-group/Beeline
- scVI https://github.com/romain-lopez/scVI-reproducibility,https://github.com/YosefLab/scvi-tools
- DCA https://github.com/theislab/dca
- ZIFA https://github.com/epierson9/ZIFA
- scGAN/cscGAN https://github.com/imsb-uke/scGAN
Some notation are incorrect in published paper.
If you have any question, please feel free to contact to me.
Email: shuht96@gmail.com
DeepSEM is licensed under the MIT License.