Hierarchical Text-to-Vision Self Supervised Alignment for Improved Histopathology Representation Learning [MICCAI 2024]
A Hierarchical Language-tied Self-Supervised (HLSS) framework for histopathology
Hasindri Watawana, Kanchana Ranasinghe, Tariq Mahmood, Muzammal Naseer, Salman Khan and Fahad Khan
Mohamed Bin Zayed University of Artificial Intelligence, Stony Brook University, Linko ̈ping University, Australian National University and Shaukat Khanum Cancer Hospital, Pakistan
- Jun-17-24: We open source the code, model, training and evaluation scripts.
- Jun-17-24: HLSS has been accepted to MICCAI 2024 🎉.
- Mar-21-24: HLSS paper is released arxiv link. 🔥🔥
Self-supervised representation learning has been highly promising for histopathology image analysis with numerous approaches leveraging their patient-slide-patch hierarchy to learn better representations. In this paper, we explore how the combination of domain specific natural language information with such hierarchical visual representations can benefit rich representation learning for medical image tasks. Building on automated language description generation for features visible in histopathology images, we present a novel language-tied self-supervised learning framework, Hierarchical Language-tied Self-Supervision (\modelname) for histopathology images. We explore contrastive objectives and granular language description based text alignment at multiple hierarchies to inject language modality information into the visual representations. Our resulting model achieves state-of-the-art performance on two medical imaging benchmarks, OpenSRH and TCGA datasets. Our framework also provides better interpretability with our language aligned representation space.
- Clone HLSS github repo
git clone https://github.com/Hasindri/HLSS.git - Create conda environment
conda create -n hlss python=3.8 - Activate conda environment
conda activate hlss - Install package and dependencies
pip install -r requirements.txt
HLSS/
├── hlss/ # all code for HLSS related experiments
├── config/ # Configuration files used for training and evaluation
│ ├── datasets/ # PyTorch datasets to work with OpenSRH and TCGA
│ ├── losses/ # HiDisc loss functions with contrastive learning
│ ├── models/ # PyTorch models for training and evaluation
│ ├── wandb_scripts/ # Training and evaluation scripts
├── figures/ # Figures in the README file
├── README.md
├── setup.py # Setup file including list of dependencies
├── LICENSE # MIT license for the repo
└── THIRD_PARTY # License information for third party code
The code base is written using PyTorch Lightning, with custom network and datasets for OpenSRH and TCGA.
To train HLSS on the OpenSRH dataset:
- Download OpenSRH - request data here.
- Update the sample config file in
config/train_hlss_attr3levels.yamlwith desired configurations. - Activate the conda virtual environment.
- Use
train_hlss_KL.pyto start training:python train_hlss_KL.py -c=config/train_hlss_attr3levels.yaml
To evaluate with your saved checkpoints and save metrics to a CSV file:
- Update the sample config file in
config/eval_hlss_attr128_accplot.yamlwith the checkpoint directory path and other desired configurations. - Activate the conda virtual environment.
- Use
eval_knn_hlss_accplot.pyfor knn evaluation:python eval_knn_hlss_accplot.py -c=config/eval_hlss_attr128_accplot.yaml
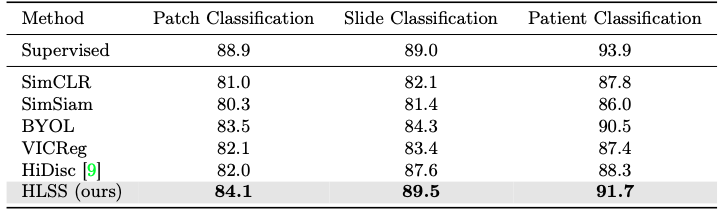
Our approach learns better representations of histopathology images from OpenSRH and TCGA datasets, as shown by the improved performance in kNN evaluation.
Should you have any questions, please create an issue in this repository or contact hasindri.watawana@mbzuai.ac.ae
Our code is build on the repository of HiDisc. We thank them for releasing their code.
If you find our work or this repository useful, please consider giving a star ⭐ and citation.
@misc{watawana2024hierarchical,
title={Hierarchical Text-to-Vision Self Supervised Alignment for Improved Histopathology Representation Learning},
author={Hasindri Watawana and Kanchana Ranasinghe and Tariq Mahmood and Muzammal Naseer and Salman Khan and Fahad Shahbaz Khan},
year={2024},
eprint={2403.14616},
archivePrefix={arXiv},
primaryClass={cs.CV}
}