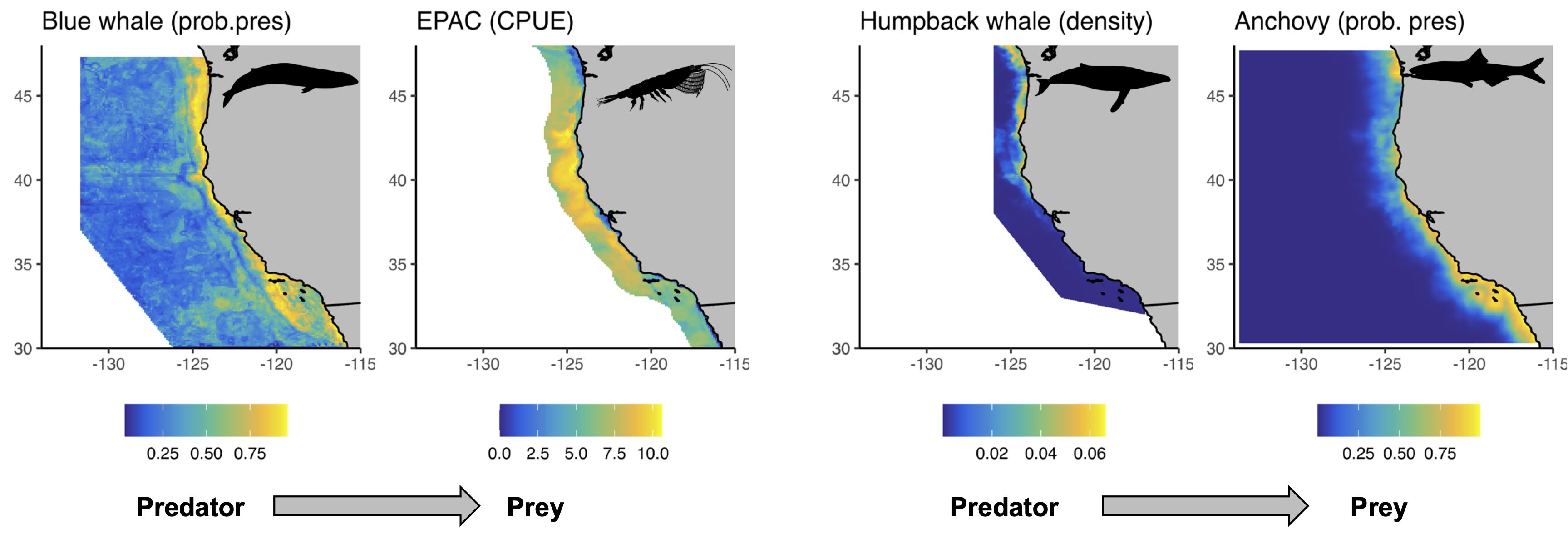
Operational predictions of blue whale (probability of presence), humpback (density), anchovy (probability of presence), and EPAC (catch per unit effort, CPUE) distributions.
For each species, outputs include:
Daily rasters of predicted distribution (.grd and .gri; "rasters")
Daily maps of predicted distribution (.png; "maps")
Daily metadata (.csv; "epac_metadata") for each EPAC prediction documenting how many days of data were available to make each rolling average monthly prediction in 1. & 2.
Rolling two week average rasters (.grd and .gri; "two_week_rasters") of predicted distribution
Rolling two week average maps (.png; "two_week_maps") of predicted distribution
Rolling two week metadata (.csv; "two_week_metadata") for each output in 4. & 5. documenting how many days of data were available. Metadata can be matched to two week predictions by “end20XX-XX-XX”
Daily indicators (.csv and .png; "ramp_zone_indicators") of average predicted distribution in RAMP zones 3 and 4
A. Navigate to the folder you want to download and copy URL, e.g. raster data for November 2023: https://github.com/HeatherWelch/OPC_Farallons_end_products_final/tree/main/rasters/2023/11
B. Paste URL in box here: https://download-directory.github.io/
C. Folder will be compressed and downloaded
A. Clone the repository: https://docs.github.com/en/repositories/creating-and-managing-repositories/cloning-a-repository
(And want to integrate products into an operational workflow)
library(RCurl)
library(tidyverse)
library(glue)
download_git=function(username,repository_name,file_path,save_dir){
save_name=file_path %>%
str_split(.,"/") %>%
unlist() %>%
.[length(.)]
git_url=glue("https://raw.githubusercontent.com/{username}/{repository_name}/main/{file_path}")
file = glue("{save_dir}/{save_name}")
f = CFILE(file,mode="wb")
a=curlPerform(url=git_url,writedata=f@ref,noprogress=FALSE, .opts = RCurl::curlOptions(ssl.verifypeer=FALSE))
close(f)
}
username="HeatherWelch"
repository_name="OPC_Farallons_end_products_final"
save_dir=getwd()
file_path="epac_metadata/2023/04/metadata_2023-04-15.csv"
download_git(username = username,repository_name = repository_name, file_path = file_path, save_dir = save_dir)
A. Krill: Cimino et al. 2020. Ecography
B. Anchovy: Fennie et al. 2023. Proc.B
C. Blue whale: Abrahms et al. 2019. Diversity and Distributions
D. Humpback whale: Unpublished. Examples of use:
Samhouri et al. 2021. Proc. B
Welch et al. 2023. Conservation Biology
https://github.com/HeatherWelch/OPC_Farallons/tree/main/Operationalizing_V2