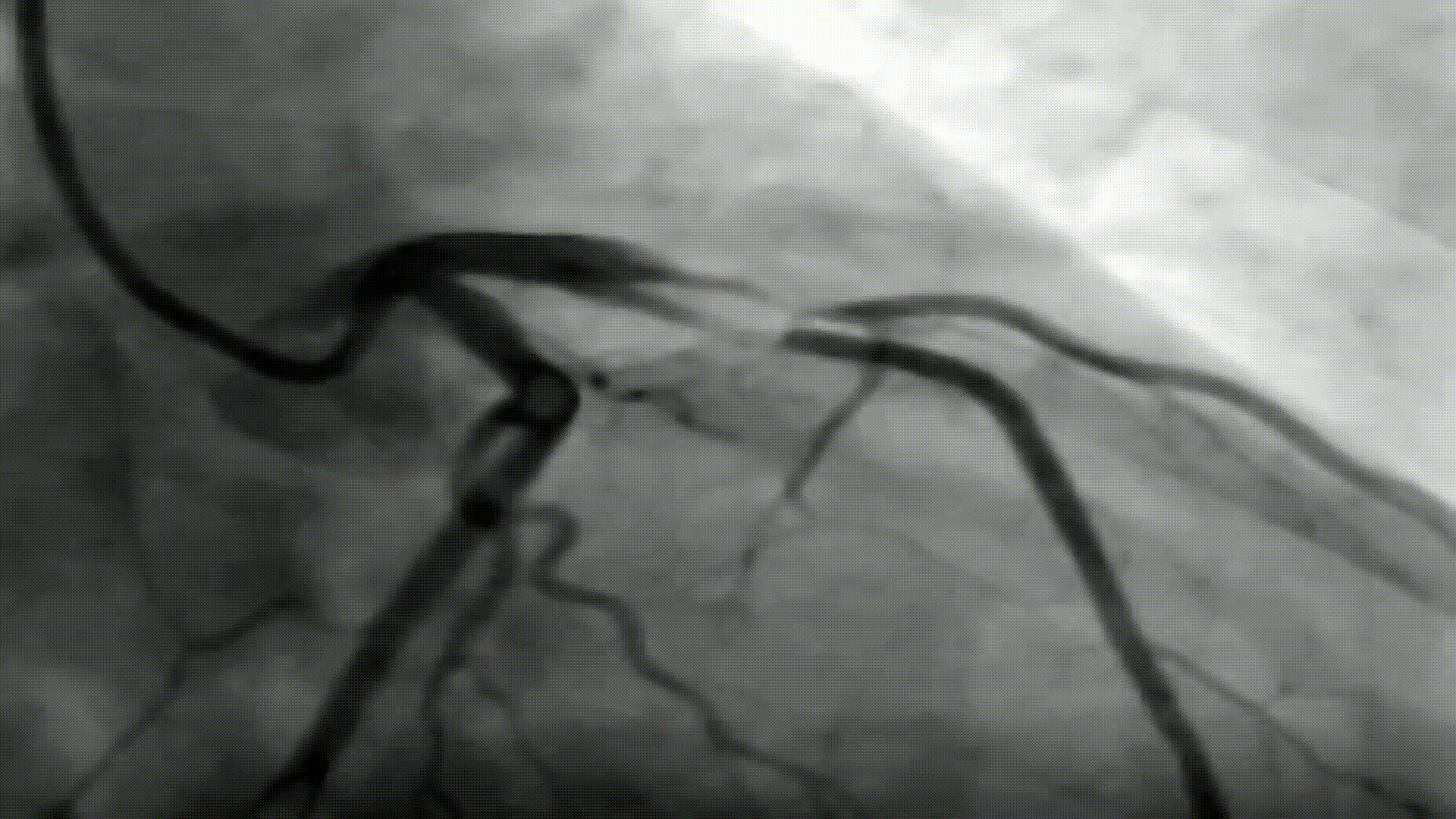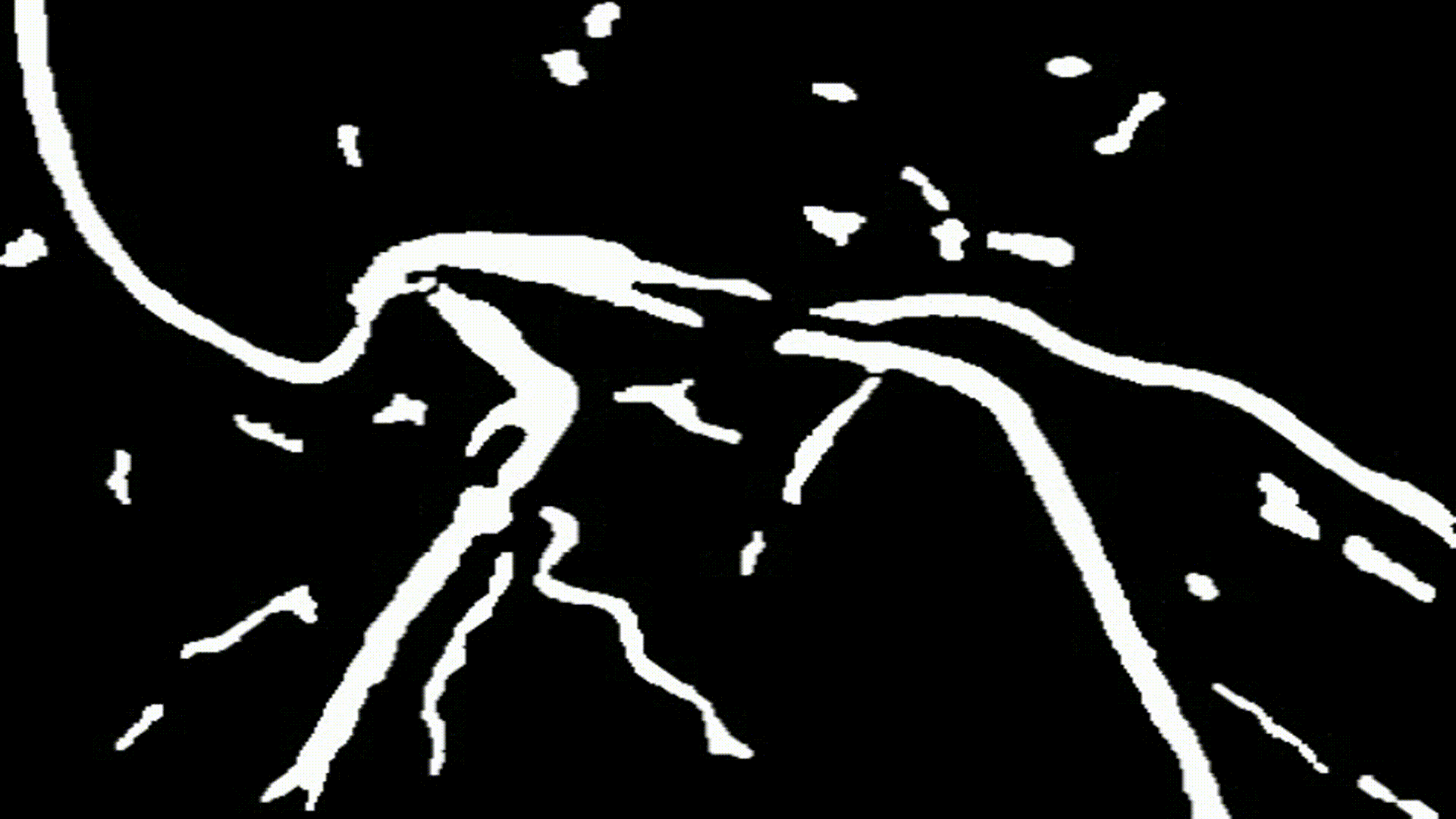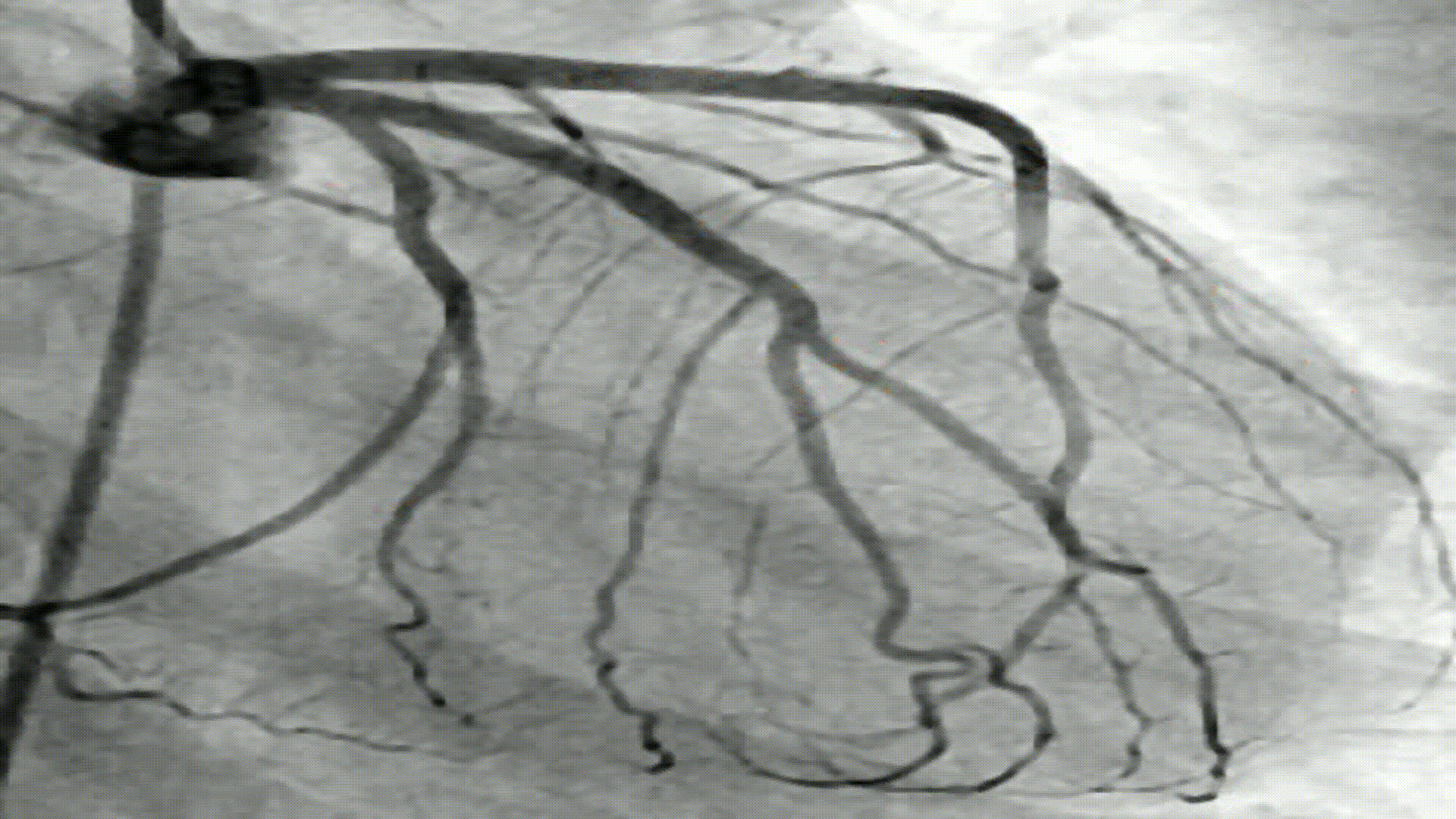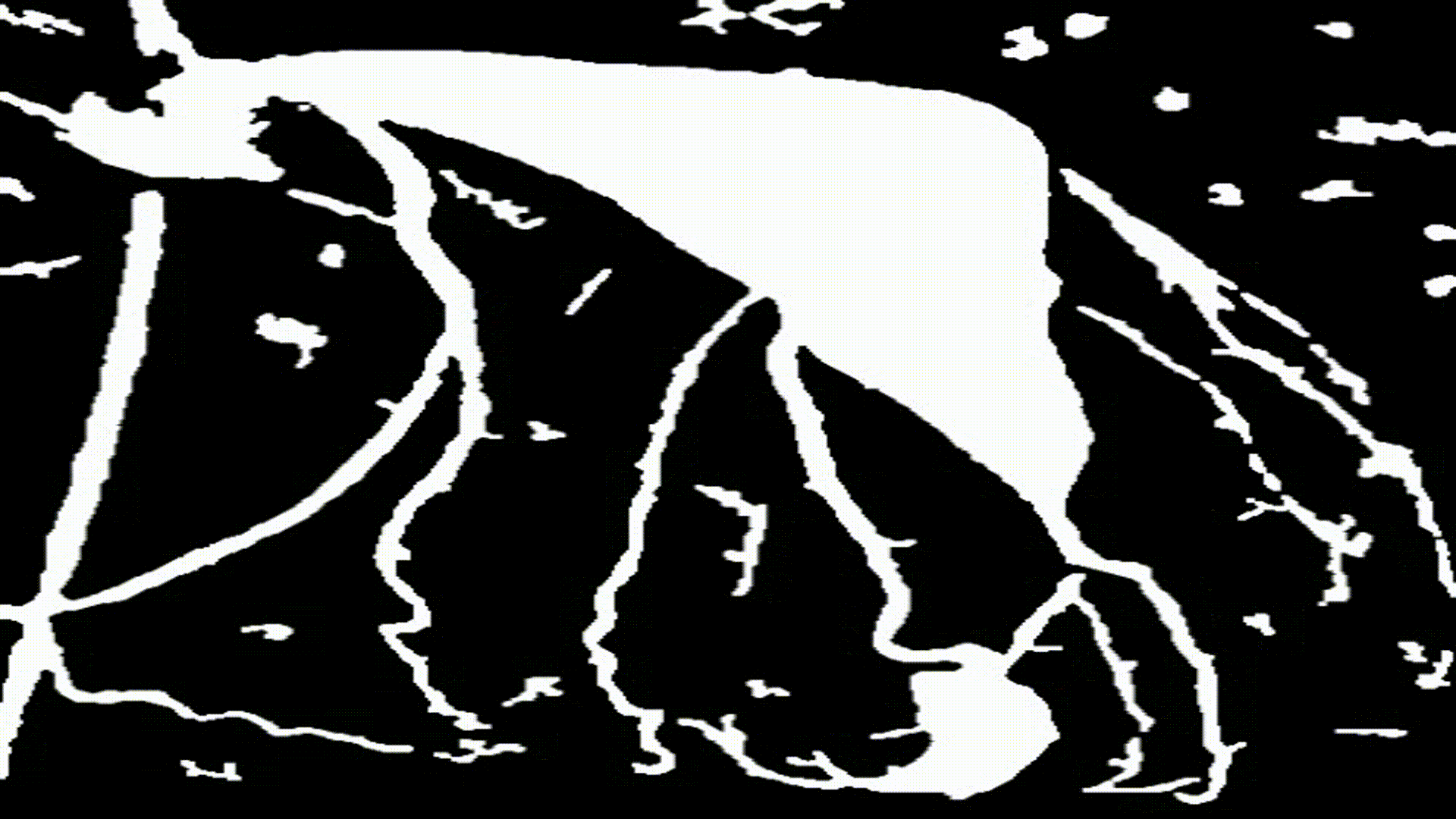
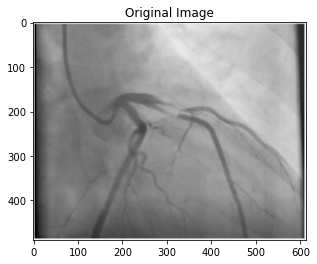
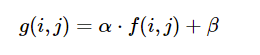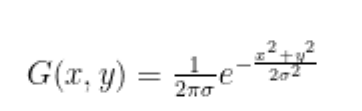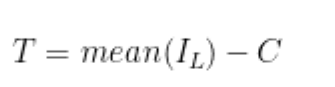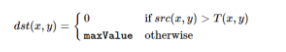Image segmentation has been one of the classical image processing techniques. It intends to abstract some important feature from an image based on its global and/or local amplitude distribution. One of the major application areas of this technique is medical imaging, where segmentation can automatically highlight the objects (e.g. vessel, cardiac chamber etc.) to help physicians diagnosing disease.
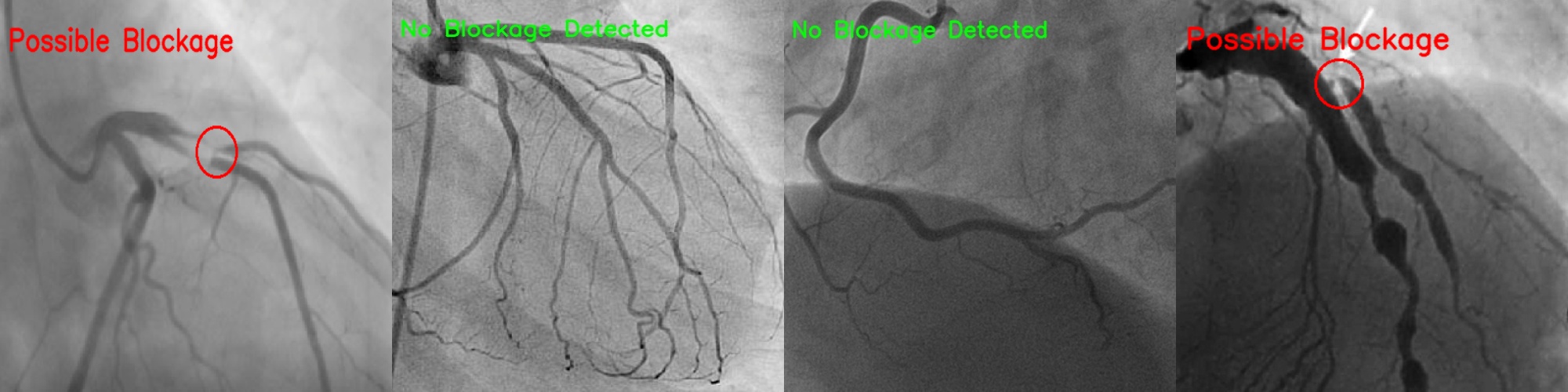
In this project, you are to apply image segmentation techniques to X-ray angiography, where X-ray images are taken when an X-ray absorbing substance is injected into the patient's blood stream to produce contrast. The resulting X-ray images have dark regions representing the blood flow within vessels. Your system should be able to automatically locate any occlusion and follow the surrounding vessel wall to compute the ratio between the minimum and nominal vessel diameters. Such results are practically important in detecting coronary disease. Your system should also accept user input of occlusion locations and perform the same percent occlusion measurement in that particular area.
- Liang, Dongxue, et al. “Coronary Angiography Video Segmentation Method for Assisting Cardiovascular Disease Interventional Treatment.” BMC Medical Imaging, vol. 20, no. 1, 2020, doi:10.1186/s12880-020-00460-9.
- Dehkordi, Maryam Taghizadeh, et al. “Retraction: A Review of Coronary Vessel Segmentation Algorithms.” Journal of Medical Signals & Sensors, vol. 9, no. 1, 2019, p. 76., doi:10.4103/2228-7477.253755.
- Sharma, Neeraj, et al. “Automated Medical Image Segmentation Techniques.” Journal of Medical Physics, vol. 35, no. 1, 2010, p. 3., doi:10.4103/0971-6203.58777.
A coronary angiogram is a low-risk medical technique used to visualize blood vessels and arteries, and can be used to both diagnose and treat heart and blood vessel conditions. To conduct this procedure, a doctor first injects a dye into a patient’s blood vessel then takes many X-ray images of this same area. Because the dye is visible in an X-ray, the doctor can determine if there is blockage (cardiovascular disease) based on the occlusion seen throughout the blood vessels. My goal for this project is to automate the process of examining these X-ray images by using a light-weight segmentation algorithm that can locate these areas of occlusion.
In the current medical industry, segmentation is a popular procedure for processing medical imagery. These methods are used to provide computer-aided diagnosis (CAD) and locate possible regions of interest (ROIs) that would be important for examining a patient’s condition and/or symptoms. However, the types of segmentation algorithms differ depending on their use case as each imaging system has its own limitations and specifications. For example, segmentation is found in common procedures such as MRIs, CT, and PET scans which use segmentation for performing amplitude segmentation based on histogram features, edge based segmentation, and region based segmentation (Sharma, 2010). Typical segmentation methods utilize a threshold that is used to compare to each pixel. If a given pixel is lower than the threshold, that pixel is converted to 0 (black) while if it is higher, it is converted to 255 (white). This can be performed in the opposite direction at the discretion of the operator.
To follow the code process, see technical jupyter notebook The process consists of 10 steps
- In X-ray images, there often can be black/white borders around the image that would negatively impact this segmentation algorithm.
- This will ensure that the distributions among pixel amplitudes is normalized.
- Using the formula below (
cv2.convertScaleAbs()), one has the ability to normalize the distributions of pixel amplitudes across an image givenalphaandbetaparameters.
-
cv2.convertScaleAbsworks by performing three operations sequentially. (1) Scaling, (2) Taking an absolute value, (3) Converting to unsigned 8-bit integer. Each new pixel is the result of performingabs(alpha + Pixel(x,y) + beta -
The function
_automatic_brightness_and_contrast(image, clip_hist_percent)found in program.py allows for dynamic calculations ofalphaandbetato perform this normalization differently on each image. This function works by:
-
Calculating the cumulative distribution of an image histogram to determine where color frequency is less than some pre-defined threshold
clip_hist_percent. -
Cut the right and left side of this histogram to give us our minimum and maximum ranges
-
Calculate
alpha = 255 / (maximum_pixel - minimum_pixel) -
Calculate beta. Given that
g(i, j) = 0andf(i, j) = minimum_pixelg(i,j) = alpha * f(i,j) + beta #0 = alpha * min_pixel + betabeta = -minimum_pixel * alpha1
-
This step will reduce noise around the image and allow for better segmentation
-
Median Blur is a commonly used method for reducing salt and pepper noise.
-
To apply a median blur, you first determine a kernel size. In this project, I used a kernel size of 5. The kernel size is used to determine the number of neighbors that will be incorporated in the blur method.
cv2.medianBlur(image, ksize = 5)
-
For example, with a neighborhood size of 5x5, 25 pixels will be used to calculate the median of all pixels.
-
After gathering the median value, the center pixel in that 5x5 area will become that median value.
-
The reason this method is so useful for salt and pepper noise is because the center pixel will always be replaced with a pixel that is in the original image- as using the median calculation is more robust to outliers than compared to using an average or gaussian blur method.
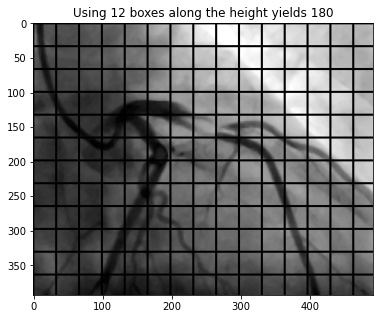
- This process involves separating the image into equal partitions so that there are a specific number of blocks along the y-axis. In this project, I ensured that there are 12 blocks along the height of the image.
- Because each block is created around a central pixel, the blockSize of an image must be an odd number and each block must have square dimensions.
- For example, if we want 12 blocks along the y-axis in an image with a dimension of 491x393 (WxH) pixels, it will require each block to have a height and width of 33 pixels. Given the height/width of each block, we can divide the width of the image by 33 to determine how many boxes will fit along the x-axis.
height_boxes = 12
block_size = int(image_height / height_boxes) #int(393 / 12) = 32 (rounded down)
if block_size % 2 == 0: #this means it is even
block_size += 1 #block size = 33
width_boxes = int(image_width / block_size) #int(491 / 33) = 14 (rounded down)
if width_boxes % 2 == 0: #this means it is even
width_boxes += 1
- The calculation used in program.py is dynamic, so these numbers are specific to the size of the image being processed
- This blockSize is utilized when applying adaptive threshold, where an algorithm will determine the best threshold based on values calculated within each of these blocks. (Explanation for how this algorithm works is explained in the next step).
- Here, I applied mean adaptive thresholding using a blockSize of 33 and a constant of 10
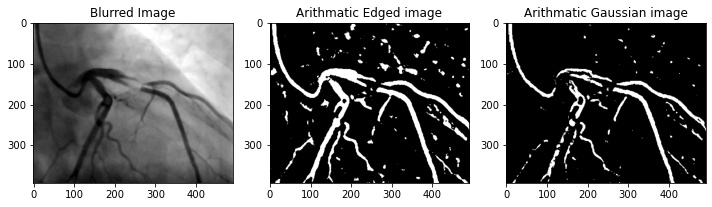
- When applying adaptive thresholding, you have the option of using Arithmetic or Gaussian mean for calculating the threshold within each image. In this project, I used Arithmetic mean (
cv2.ADAPTIVE_THRESH_MEAN_C) as I believe Gaussian mean is not a good method for this application. In Gaussian mean (cv2.ADAPTIVE_THRESH_GAUSSIAN_C), the weighted average is performed so that the central pixel of the block contributes more weight to the average. In the example image below, we can see that Gaussian mean reduces noise present in the image, however, it does not preserve the integrity of the vessels as well as arithmetic mean.
cv.ADAPTIVE_THRESH_MEAN_C: The threshold value is the mean of the neighbourhood area minus the constant C.
cv.ADAPTIVE_THRESH_GAUSSIAN_C: The threshold value is a gaussian-weighted sum of the neighbourhood values minus the constant C.
- The threshold for each block is calculated by taking the arithmetic mean of the (blockSizexBlockSize) and subtracting it by
C = 10. In the example in the previous step, using 12 block rows yields a blockSize of 33. Given this, we will take the arithmetic average pixel amplitude within each 33x33 block and subtract that average by 10 to determine the threshold for that specific block. As mentioned in Step 6, the blockSize will change based on the original image dimension so the math explained here applies only to that image - however, the logic is the same. In Step 6, I obtained 12 block rows (y) and 15 block columns (x). Therefore, there will be a total of 180 (12 * 15) thresholds that correspond to each partitioned area.
- After determining the thresholds for each specific block, an algorithm is applied where each pixel in a particular block is converted to either 0 (black) or 255 (white) based on that block's calculated threshold. There are 2 procedures that are popular:
cv2.THRESH_BINARY: Each pixel greater than or equal to the threshold value will be converted to a defined max value (255) while every pixel below the threshold will be converted to 0 (black)
cv2.THRESH_BINARY_INV: Each pixel greater than or equal to the threshold value will be converted to 0 (black) while every pixel below the threshold will be converted to a defined max value (255)
- In this project, I used
cv2.THRESH_BINARY_INVwhich converted every pixel above the threshold to black while converting pixels below the threshold to white (max value of 255). By doing this, we can interpret the white regions in the image as possible blood cells and thus our regions of interest.
threshold_img = cv2.adaptiveThreshold(original_image, 255, cv2.ADAPTIVE_THRESH_MEAN_C, cv2.THRESH_BINARY_INV, block_size, 10)
- I performed a contour operation to find the edges within the image. These edges are the white areas shown in Step 7
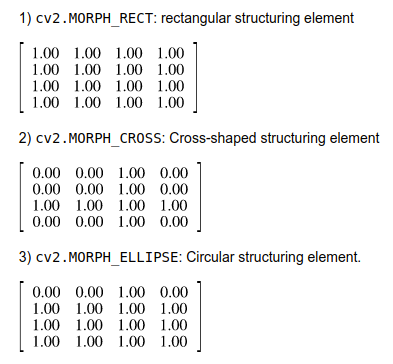
- In order to perform a contour operation, I first created a Structuring Element: which is a type of kernel that performs a particular operation on the image.
- When creating a structuring element, popular morphological operations are
-
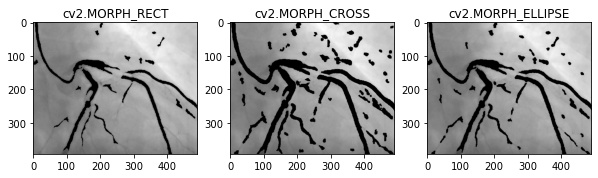
In this project, I used circular structuring element (
cv2.MORPH_CROSS) and a kernel size of (3x3). In the image below, the circular structuring element was better at maintaining the blood vessel. -
Using
cv2.morphologyEx, I apply a convolution between each 3x3 block in the image with the ELLIPSE_MATRICS above matrix. -
After convolving the image, we apply
cv2.findContourswithcv2.RETR_EXTERNALwhich is an algorithm that detects changes in colors and perceives them as boundaries. In this case, every point where a black pixel is right next to a white pixel is processed as a boundary. This function will return an array of values, where the length of the array corresponds to each contours region. I parsed the array to only include contours with a minimum area of 70 pixels -
In the image below, the contour regions are those drawn in black. Although the image looks similar to the ones in previous steps, the difference here is that I obtained (x,y) coordinates for each point around the blood vessels.
- Given the images created in Step 8, I applied another threshold operation to create a binary image where every pixel is either 0 (black) or 255 (white)
- Because I drew and filled each contour in black, I know that the blood vessels have a pixel value of 0
- I apply a simple threshold algorithm that will turn every pure black pixel to 255, while converting any other pixel greater than 0 to 255.
- Dilation (
cv2.dilate()) Is the process of that increases the bright regions of the image. The process of dilation is as follows
- Create a kernel and scan the image with that kernel
- Within each overlapping block of the kernel and the original image, we replace the center pixel with the maximum value found in that block.
- If more than one iteration is passed, then you repeat this process for the remaining iterations. The more iterations you pass, the brighter the image will get.
- As shown in the image below, performing dilation with
iterations = 2on this image decreased the minimum distance between each contour point, allowing for better blockage estimation in the final step.
- Here, we perform the operations as step 8 on the dilated image retrieved from step 7.
- We filter the array of contours to include only contours with a minimum shape area of 1250 pixels so that we limit the amount of calculations and also avoid performing calculations on artifacts.
- Once we have the contour points of the dilated image, we apply a contour euclidean distance formula _find_if_close to get the distance of each contour to all other contours.
- The algorithm for finding the euclidean distance operates using brute force iterations. For future iterations of this project, I plan on implementing this using broadcasting which would increase the speed of this calculation.
- If the distance between two different contours is less than or equal to
min_dist = 10, then we can assume that is a blockage area of the blood vessel.
- Clone repository onto your local machine
git clone <repo url>
- Set up virtual environment (conda, etc.) [Recommended]
- Install Miniconda
cd /tmpapt-get update && apt-get install wget -y && wget https://repo.anaconda.com/miniconda/Miniconda3-py39_4.10.3-Linux-x86_64.shchmod +x Miniconda3-py39_4.10.3-Linux-x86_64.sh && ./Miniconda3-py39_4.10.3-Linux-x86_64.sh- Follow the steps prompted in the command line
- Restart terminal to initialize conda
- You should now see
(base)on the left of your new terminal session
- You should now see
- Create conda environment
conda create --name blockage-detection python=3.9conda activate blockage-detection- You should now see that
(base)has changed toblockage-detection
- You should now see that
- Install Miniconda
- Install necessary packages
- [conda]
pip install opencv-contrib-python matplotlib tqdm && conda install pyqt - [no virtual enviornment]
pip3 install opencv-contrib-python matplotlib tqdm PyQT5
- [conda]
- Gather images you wish to run inference on and place them in sample-images
- Run the program
- [conda]
python program.py - [no virtual environment]
python3 program.py
- [conda]
- Upon running program.py a command line prompt will get displayed asking if you wish to specify a region of interest
- This step is to determine if you want the algorithm to process the entire image or just a region of interest that you get to draw
- Press Y to draw the region of interest | Press N to let the algorithm process the entire image
- If you pressed Y, a window will pop up displaying one of your images in the sample-images directory
- Once the application is done iterating through the images inside sample-images, the output images will be saved in output-images
- The default configuration will save only the output image with the detection, however, if you want to every step of the process, change
save_all_stepson the bottom of program.py toTrue
- The default configuration will save only the output image with the detection, however, if you want to every step of the process, change
- Once the program is done processing each image, it will display the detections in another window. Upon closing this window, the output-image and steps taken for each original image will be saved in output-images
ImportError: libGL.so.1: cannot open shared object file: No such file or directory- This is an opencv package conflict, in order to fix it you must run
apt-get update && apt-get install ffmpeg libsm6 libxext6 -y
- This is an opencv package conflict, in order to fix it you must run
- Python version
- To check your python version, open a command line and run
python- If the version is < 3.0.0, then you must use
python3to run the program - run
python3 program.pyinstead ofpython program.py
- If the version is < 3.0.0, then you must use
- To check your python version, open a command line and run
no protocol specified qt.qpa.xcb: could not connect to display :1 qt.qpa.plugin: Could not load the Qt platform plugin "xcb" in "/root/miniconda3/envs/blockage-detection/lib/python3.9/site-packages/cv2/qt/plugins" even though it was found. This application failed to start because no Qt platform plugin could be initialized. Reinstalling the application may fix this problem.- This means you do not have pyqt installed.
- If you are running conda outside of the container,
conda install pyqt - If you are not running a virtual environment,
pip3 install PyQt5
- If you are stuck on a different error and need assistance, create an issue on this repo and I will be sure to answer ASAP
- experiment ridge detection, piece-wise normalization, Harris corner detection and the rolling algorithm
- use segmentation networks and other machine learning approached to generate similar or better results
- make the function _find_if_close for calculating the euclidean distance between two contours faster
- Test with images that are smaller than (500,500) without having to resize them to larger dimensions