Fully automatic computer-aided mass detection and segmentation via pseudo-color mammograms and Mask R-CNN
Algorithm for simultaneous detection and segmentation of mammographic masses via pseudo-color mammograms and Mask R-CNN.
-
Code: [GitHub Repository – https://github.com/Holliemin9090/Mammographic-mass-CAD-via-pseudo-color-mammogram-and-Mask-R-CNN]
-
Mask R-CNN code: [GitHub Repository – https://github.com/matterport/Mask_RCNN]
-
Data: INBreast dataset
-
Trained model example: [GitHub Pages - https://github.com/Holliemin9090/Mammographic-mass-CAD-via-pseudo-color-mammogram-and-Mask-R-CNN/releases/tag/v1]
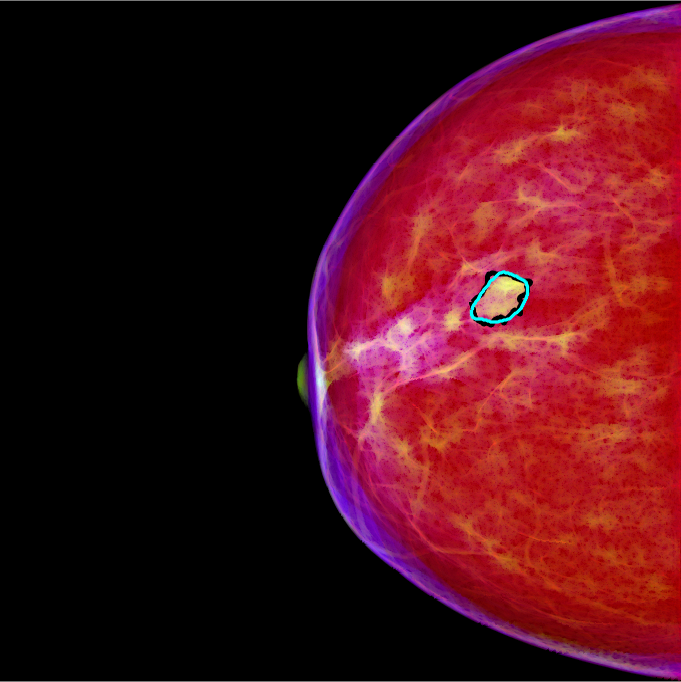
The proposed method transforms conventional grayscale mammograms into pseudo-color mammograms using multi-scale morphological sifting (MMS). The MMS can enhance lesion-like patterns within a specified size range. Two scales are used in MMS and two output images are generated. These two images are then appended to the grayscale mammogram to form a RGB pseudo-color image. The lesion-like patterns will show color contrast to the background in the new pseudo-color image as shown in the image above (the blackline represents the annotation and the cyan line represents the segmentation generated by the proposed method).
The pseudo-color mammograms are then used as inputs for the Mask R-CNN. The Mask R-CNN is trained to perform simultaneous detection and segmentation of breast masses. We notice that using pseudo-color mammograms improved the detection and segmentation performance of Mask R-CNN when compared with using conventional grayscale mammograms.
The method consists of 3 steps: pre-processing, pseudo-color image generation and Mask R-CNN detection/segmentation.
This step is to normalize the raw mammograms and pad them into a square shape.
The main script is “Preprocess_mammo_mask.py”. The normalization function is in module “Preprocess_mammo.py” and the padding function is in “utility.py”.
This step is to generate two output images by applying a 2-scale morphological sifting (MS) on the mammograms. The grayscale mammogram is put in the R (red) channel, the output from the smaller scale MS is put in the G (green) channel and the output from the larger scale is put in the B (blue) channel.
- Details on multi-scale morphological sifting (MMS): Multi-scale sifting for mammographic mass detection and segmentation
The main script is "Pseudo_color_image_generation.m".
Function "Morphological_filter_bank.m" is to generated the magnitudes of the linear structuring elements. Function "Morphological_sifter.m" is to perform MMS on the input image.
We use transfer learning with Mask R-CNN due to the size limitation of the dataset used. The pre-trained model "mask_rcnn_balloon.h5" needs to be downloaded.
Pre-trained weights : mask_rcnn_balloon.h5
The Mask_RCNN code needs to be downloaded and put in the "Mask_r_cnn" folder.
The main script for training and testing is "mamo_CAD.py". The configuration for Mask R-CNN has been specified in this file. We used 5-fold cross validation in our paper, and one of the trained models is uploaded as an example under the name of "mask_rcnn_mamogram_weights.h5".
- For training, run:
python mamo_CAD.py train --weights=mask_rcnn_balloon.h5
- For testing, run:
python mamo_CAD.py segment --weights=logs/mask_rcnn_mamogram_weights.h5 --image=scans/pseudo_color_image/
The main script is "overlay_detection_segmentation_results.m" which calculates the true positive rate (TPR), the false positive rate (FPR) and the Dice similarity index (DSI) of the segmentation.
The proposed method is implemented in MATLAB R2018a and Python 3.6.