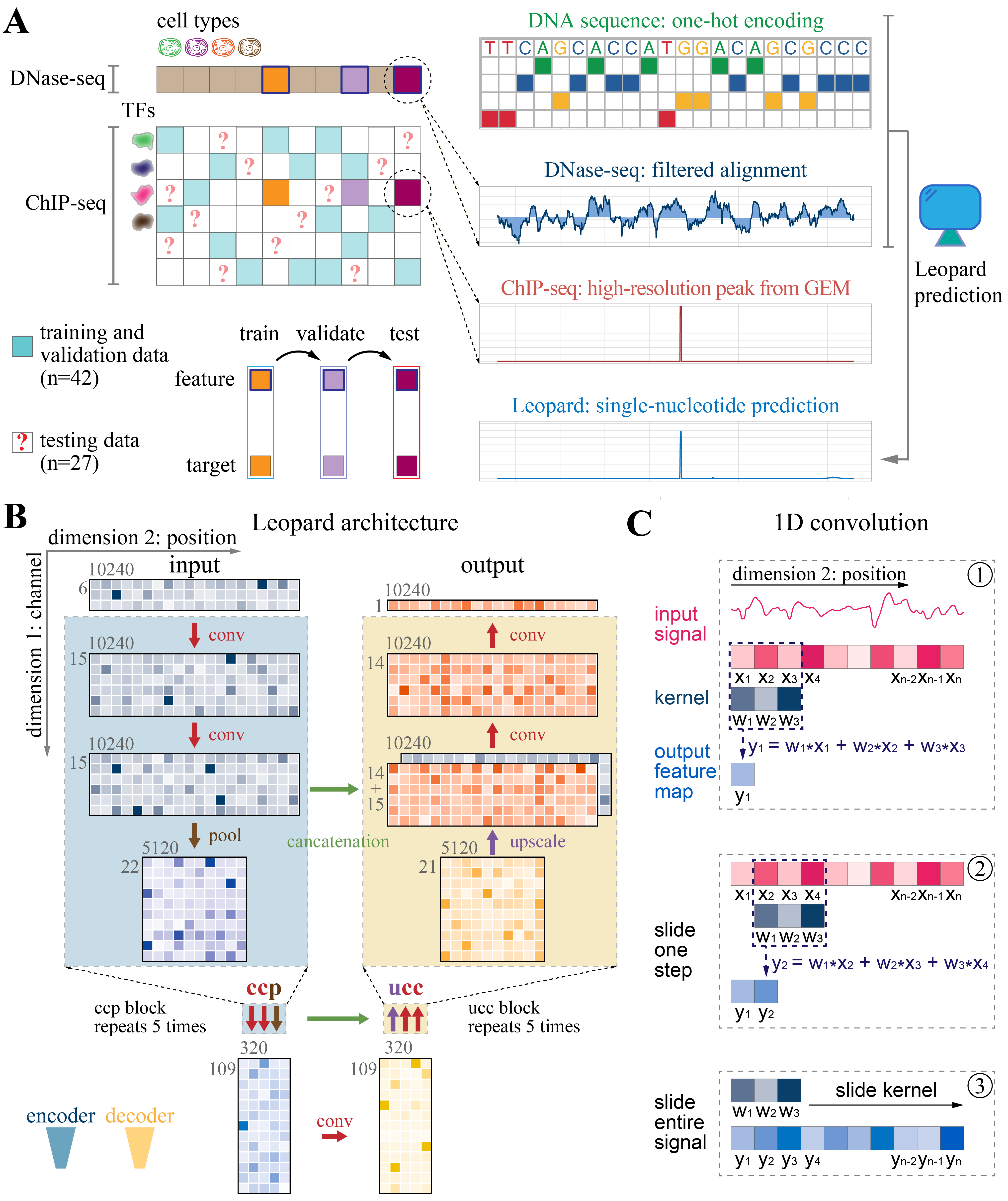
Leopard: fast decoding cell type-specific transcription factor binding landscape at single-nucleotide resolution
Leopard is a deep learning approach to predict cell type-specific in vivo transcription factor binding sites with high accuracy, speed and resolution Hongyang Li, Yuanfang Guan - Genome Research. 2021. 31: 721-731. Please contact (hyangl@umich.edu or gyuanfan@umich.edu) if you have any questions or suggestions.
Git clone a copy of code:
git clone https://github.com/GuanLab/Leopard.git
- python (3.6.5)
- numpy (1.13.3). It comes pre-packaged in Anaconda.
- pyBigWig A package for quick access to and create of bigwig files. It can be installed by:
conda install pybigwig -c bioconda
- tensorflow (1.14.0) A popular deep learning package. It can be installed by:
conda install tensorflow-gpu
- keras (2.2.5) A popular deep learning package using tensorflow backend. It can be installed by:
conda install keras
The data in bigwig format can be directly downloaded from OSF repository
- DNase-seq,DNA sequence,ChIP-seq GEM peak or from our web server:
- DNase-seq
- DNA sequence
- ChIP-seq GEM peak
Before running Leopard, please download the above data (30GB) and deposit them in the "Leopard/data/" folder. The DNA sequence bigwig files are always needed. If you only need to make predictions on one cell type, you only need to download the "avg.bigwig" and the correpsonding DNase-seq file for this specific cell type. The ChIP-seq data are optional. You only need them if you want to re-train/adapt our models or compare predictions with experimental observations.
The original data can be found as follows:
The DNase-seq data were downloaded from the ENCODE-DREAM challenge website: filtered alignment.
The ChIP-seq data were from the ENCODE project with the accession numbers provided in Supplementary Table S9.
The challenge-related ChIP-seq data were downloaded from the ENCODE-DREAM challenge website: conservative peaks and fold enrichment.
Once the required input files are put in the correpsonding directories, Leopard is ready to go (fast mode):
python Leopard.py -tf E2F1 -te K562 -chr chr21 chr22
Or you can run the complete mode with higher accuracy and longer runtime:
python Leopard.py -tf E2F1 -te K562 -chr chr21 -m complete
The prediction npy files are saved in the ./output/ folder
Here I use a new cell line, Ag04449, as an example to demontrate how to performan quantile normalization and generate the average signals for the delta-DNase-seq. The reference genome is GRCh37/hg19.
The Ag04449 has two replicates and we download both of them. It also works if you only have one replicate.
cd data
wget ftp://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeUwDnase/wgEncodeUwDnaseAg04449RawRep1.bigWig
wget ftp://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeUwDnase/wgEncodeUwDnaseAg04449RawRep2.bigWig
You can choose your own reference cell line. As long as you use the same reference cell line, it won't affect the result too much in most situations.
wget https://guanfiles.dcmb.med.umich.edu/dnase_bigwig/liver.bigwig
Subsampling a subset to estimate the overall distribution, which will save a lot of time and memory. A new file called "sample_liver.npy" will be generated in the "output" directory
python subsample_for_qn.py -i liver.bigwig -o sample_liver.npy -rg grch37
In this example, the two Ag0449 replicates are the input files. This code accepts single or multiple replicates. A new file called "sample_Ag04449.npy" will be generated in the "output" directory.
python subsample_for_qn.py -i wgEncodeUwDnaseAg04449RawRep1.bigWig wgEncodeUwDnaseAg04449RawRep2.bigWig -o sample_Ag04449.npy -rg grch37
Based on the subsampled data from the previsous two steps, quantile normalize the genome-wide signal of Ag04449 to the reference liver cell line.
python quantile_normalize_bigwig.py -r ./output/sample_liver.npy -s ./output/sample_Ag04449.npy -i wgEncodeUwDnaseAg04449RawRep1.bigWig wgEncodeUwDnaseAg04449RawRep2.bigWig -o Ag04449.bigwig -rg grch37
In Leopard, we used all 13 cell lines to calculate the avg.bigwig.Of note, when a new testing cell line comes, you don't need to re-calculate this reference and you can directly use "avg.biwig" we provided. In general,about 10 cell lines should be enough to generate a robust average signal. In case you are interested in calculating a new average, here we use liver and Ag04449 as an example. A new file called "avg_new.bigwig" will be generated.
python calculate_avg_bigwig.py -i liver.bigwig Ag04449.bigwig -o avg_new.bigwig -rg grch37