Ocelot: Improved Epigenome Imputation Reveals Asymmetric Predictive Relationships Across Histone Modifications
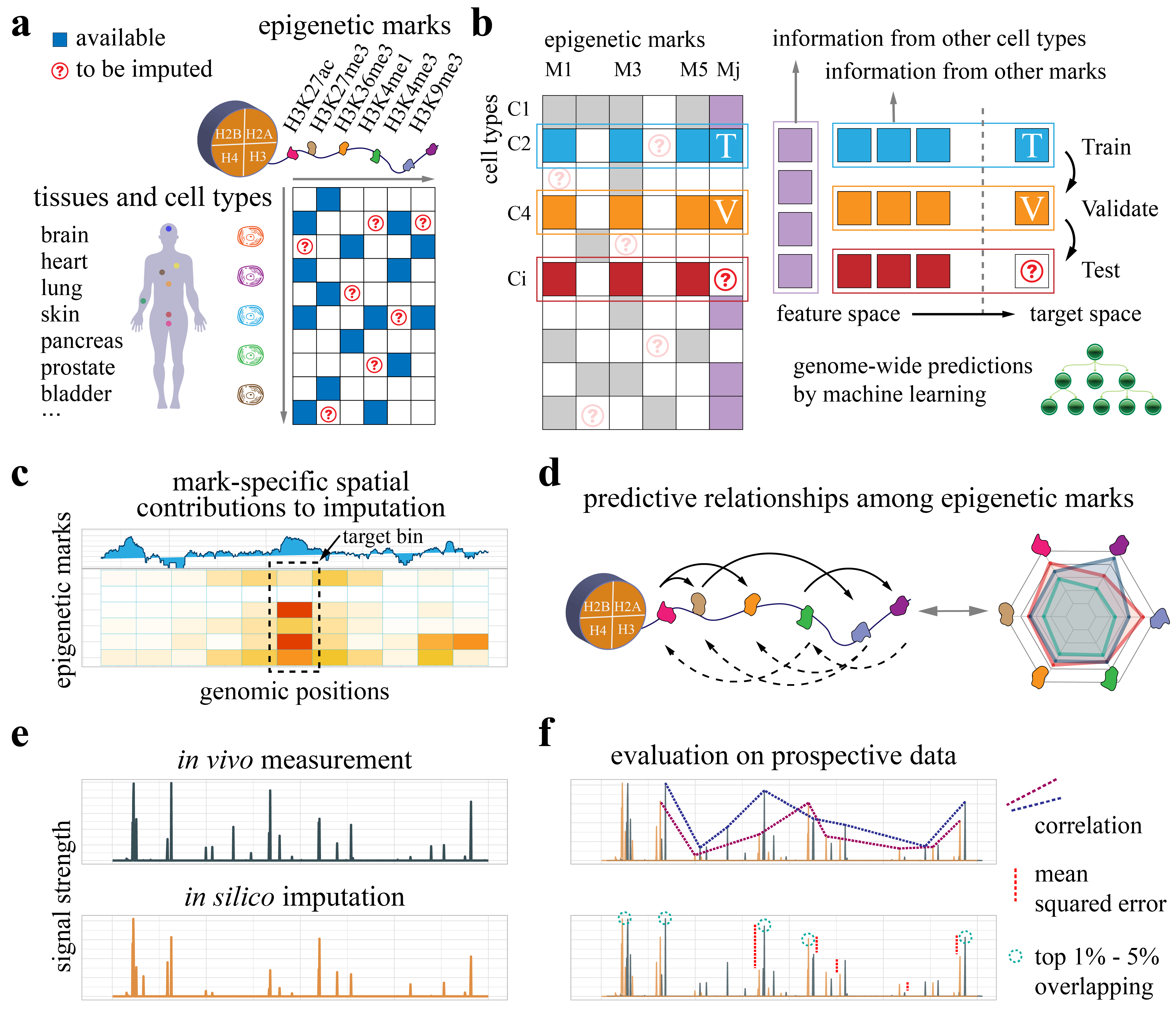
Ocelot is a machine learning approach to impute epigenomes across tissues and cell types. It ranked first in the ENCODE Imputation Challenge with high accuracy on held-out prospective data. Beyond high predictive performance, it offers a new way to investigate the cross-histone regulations based on large-scale epigenomics datasets. Please contact (hyangl@umich.edu or gyuanfan@umich.edu) if you have any questions or suggestions.
Git clone a copy of code:
git clone https://github.com/GuanLab/Ocelot.git
- python (3.6.5)
- numpy (1.13.3). It comes pre-packaged in Anaconda.
- pyBigWig A package for quick access to and create of bigwig files.
conda install pybigwig -c bioconda
- lightgbm(2.3.0) A gradient boosting tree-based algorithm with fast training speed and high efficienty.
pip install lightgbm
- tensorflow (1.14.0) A popular deep learning package.
conda install tensorflow-gpu
- keras (2.2.5) A popular deep learning package using tensorflow backend.
conda install keras
- The ENCODE Imputation Challenge dataset
- Ocelot imputation for the ENCODE3 histone mark dataset
- Ocelot imputation for the Roadmap histone mark dataset
- data_challenge
- code_challenge
Reproducing all these imputations and evaluations requires considerable time even with super computing resources, we therefore also provide the processed data, trained models and predictions together with the reproducible scripts.
- 0a. Ocelot - the challenge final submission or npy format
- 0b. Ensemble predictions without DNA
- 1a. Processed data
- 2a. Trained lightGBM and neural network models and predictions
- 2b. Trained lightGBM and neural network models without DNA and predictions
For benchmarking, predictions from Avocado and ChromImpute are also provided:
For simplicity, we map the epigeneic marks to captital letters as follows:
| letter | id | mark |
|---|---|---|
| C | M02 | DNase-seq |
| D | M18 | H3K36me3 |
| E | M17 | H3K27me3 |
| F | M16 | H3K27ac |
| G | M20 | H3K4me1 |
| H | M22 | H3K4me3 |
| I | M29 | H3K9me3 |
| J | M01 | ATAC-seq |
For example, in the "CDEH_I" design, we used four marks (C, D, E, H) as cell type-specific features to predict mark I.
- data_encode3
- code_encode3
- data_roadmap
- code_roadmap
- code_shap