This is the package of our winning algorithm in the NCI-CPTAC DREAM Proteogenomics Challenge.
background: Proteogenomics Challenge
see also: Hongyang Li and Yuanfang Guan's 1st Place Solution
Please contact (hyangl@umich.edu or gyuanfan@umich.edu) if you have any questions or suggestions.
Git clone a copy of code:
git clone https://github.com/GuanLab/proteome_prediction.git
- R (3.4.3)
- python (3.6.5)
- numpy (1.13.3). It comes pre-packaged in Anaconda.
- scikit-learn (0.19.0) A popular machine learning package. It can be installed by:
pip install -U scikit-learn
All the omic data are 2D matrices, where columns are cancer samples and rows are genes/proteins. The CNV and RNA-seq data originally came from TCGA. The proteomic data originally came from CPTAC.
We directly downloaded the data from the challenge website and more details can be found at: https://www.synapse.org/#!Synapse:syn8228304/wiki/448372
To run the code, download the following omic data from here and put them into the directory data/raw/
- retrospective_breast_CNA_sort_common_gene_16884.txt
- retrospective_breast_proteome_sort_common_gene_10005.txt
- retrospective_breast_RNA_sort_common_gene_15107.txt
- retrospective_ova_CNA_sort_common_gene_11859.txt
- retrospective_ova_JHU_proteome_sort_common_gene_7061.txt
- retrospective_ova_PNNL_proteome_sort_common_gene_7061.txt
- retrospective_ova_rna_seq_sort_common_gene_15121.txt
Then preprocess the data and generate 5-fold cross validatation using code in
- data/trimmed_set
- data/normalization
- data/cv_set
We have two sets of code in parallel
- prediction/breast
- prediction/ova
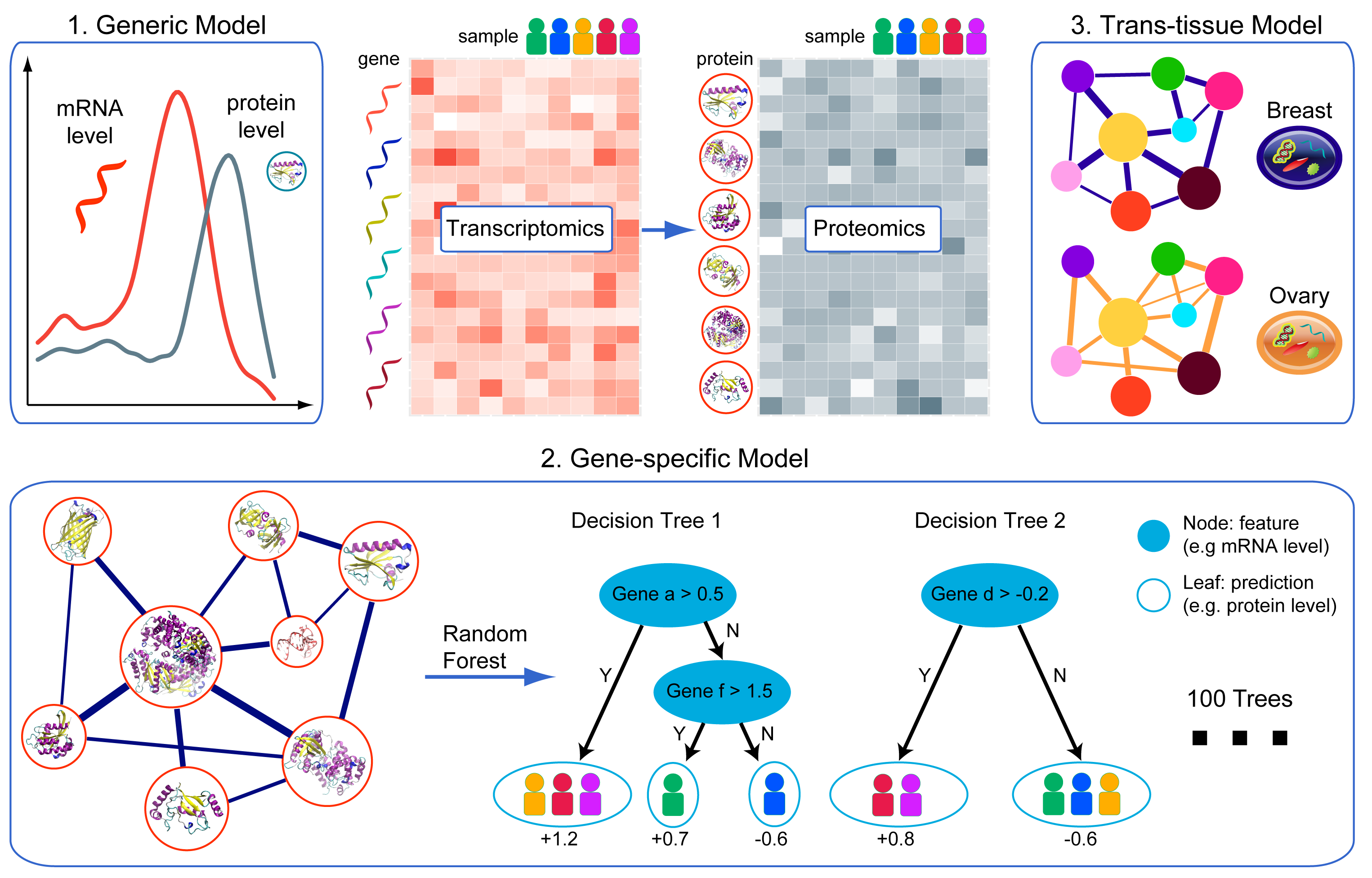
This model directly approximates the protein level based on the corresponding mRNA level.
prediction/breast/rna/
This model considers the gene-gene interactions in regulating protein abundance, in which mRNA levels of all genes were used as features to make predictions. The base learner is random forest with maximum depth of 3 and 100 trees.
prediction/breast/individual/
Similar to the gene-specific model, this model uses combined samples from breast and ovarian cancer samples.
prediction/breast/individual_transplant/
Our final results are the ensemble of the 1-3 models mentioned above.
prediction/breast/final/
analysis/