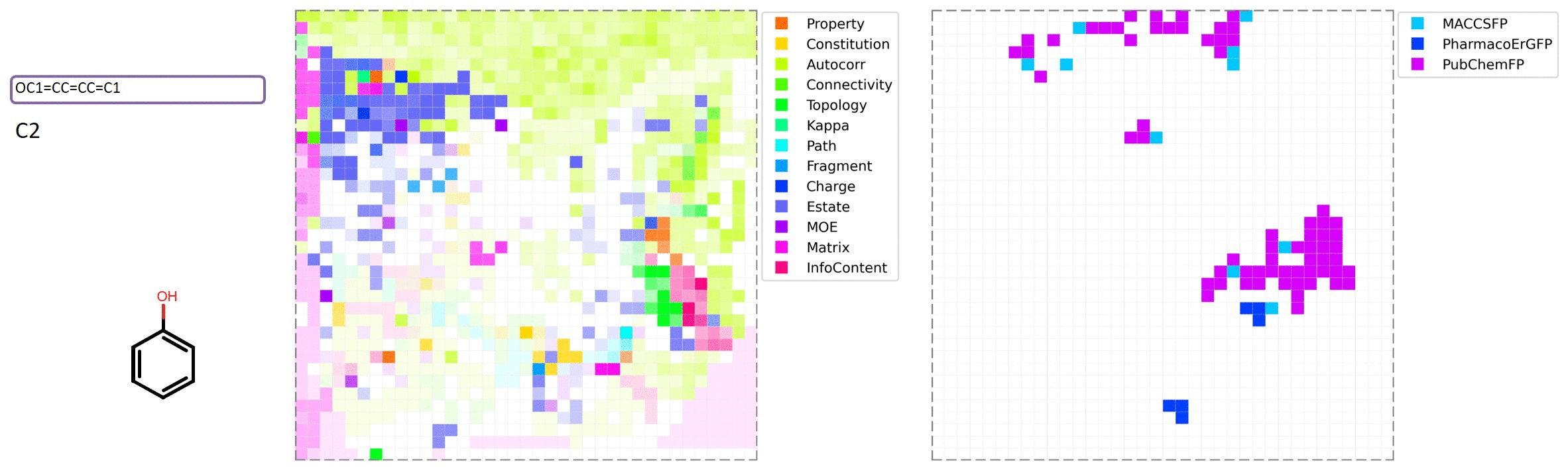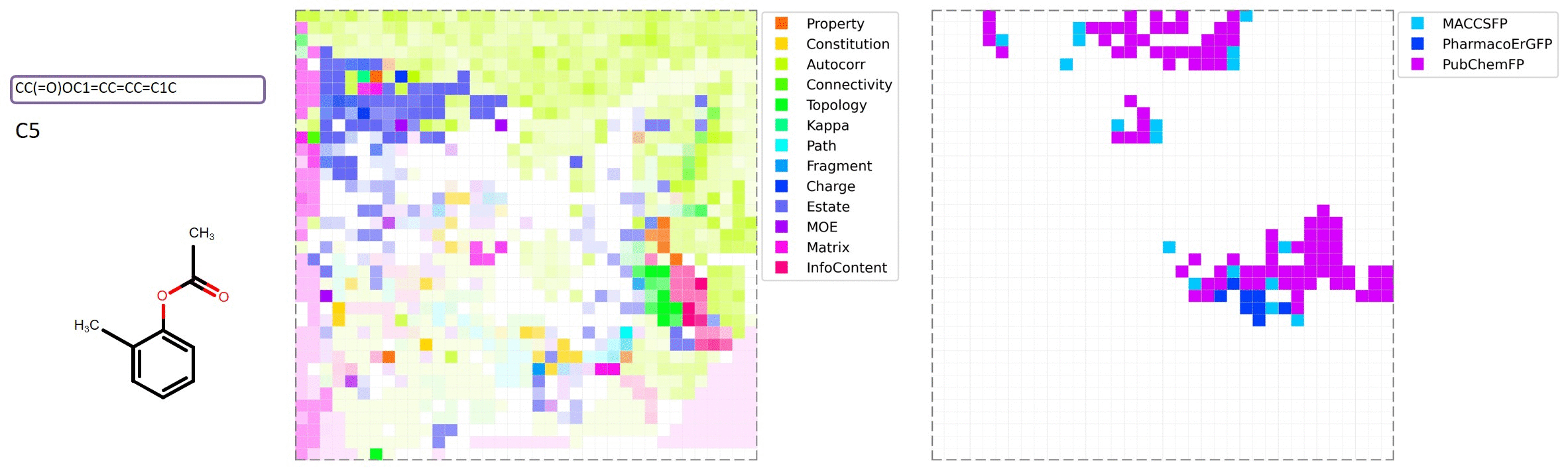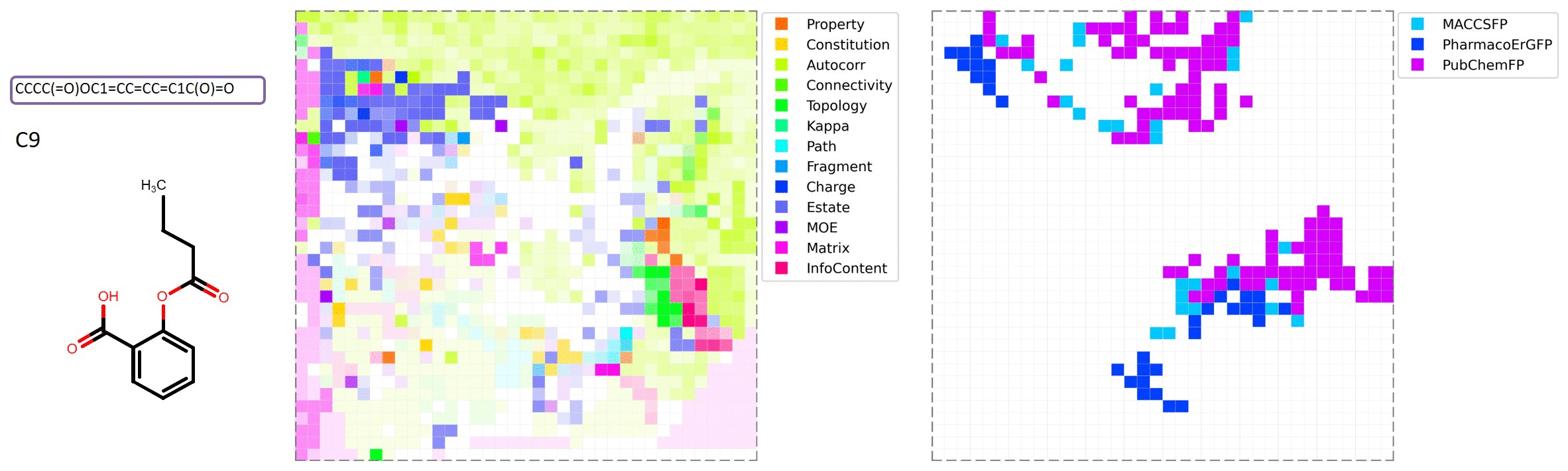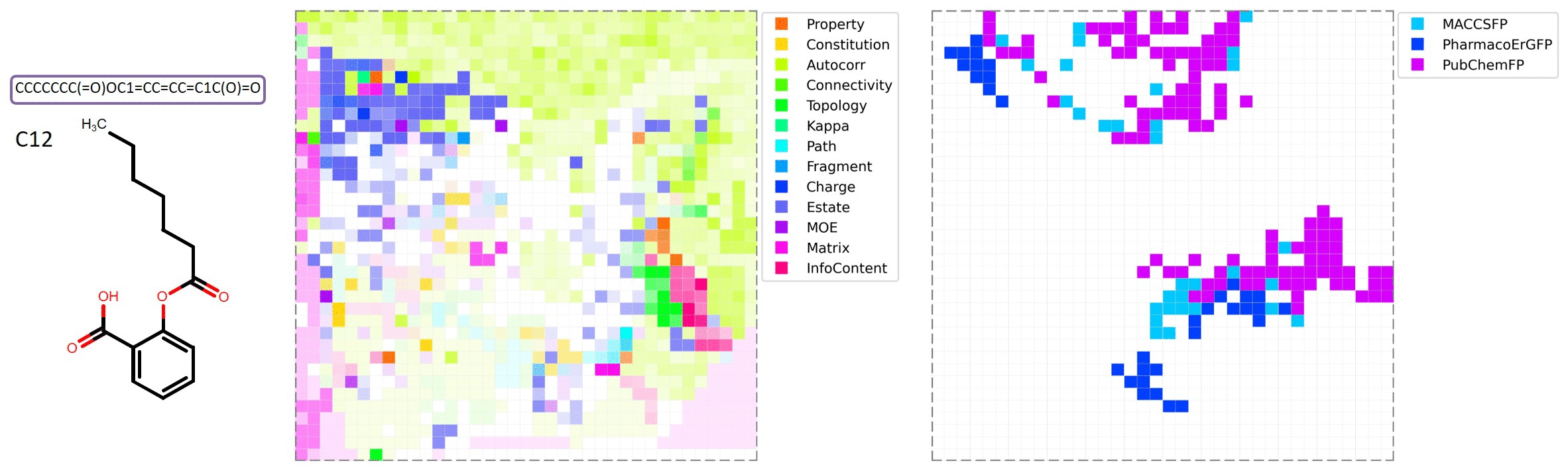
MolMap is generated by the following steps:
- Step1: Input structures
- Step2: Feature extraction
- Step3: Feature pairwise distance calculation --> cosine, correlation, jaccard
- Step4: Feature 2D embedding --> umap, tsne, mds
- Step5: Feature grid arrangement --> grid, scatter
- Step5: Transform --> minmax, standard
conda create -c conda-forge -n molmap rdkit python=3.7
conda activate molmap
conda install -c tmap tmap
pip install molmap-
ChemBench (optional, if you wish to use the dataset and the split induces in this paper).
-
If you have gcc problems when you install molmap, please installing g++ first:
sudo apt-get install g++-
Example for Regression Task on FreeSolv (descriptors plus fingerprints)
-
Example for Classification Task on BACE (descriptors plus fingerprints)
-
Example for Multi-label Classification Task on ClinTox (descriptors plus fingerprints)
import molmap
# Define your molmap
mp_name = './descriptor.mp'
mp = molmap.MolMap(ftype = 'descriptor', fmap_type = 'grid',
split_channels = True, metric='cosine', var_thr=1e-4)# Fit your molmap
mp.fit(method = 'umap', verbose = 2)
mp.save(mp_name) # Visulization of your molmap
mp.plot_scatter()
mp.plot_grid()# Batch transform
from molmap import dataset
data = dataset.load_ESOL()
smiles_list = data.x # list of smiles strings
X = mp.batch_transform(smiles_list, scale = True,
scale_method = 'minmax', n_jobs=8)
Y = data.y
print(X.shape)# Train on your data and test on the external test set
from molmap.model import RegressionEstimator
from sklearn.utils import shuffle
import numpy as np
import pandas as pd
def Rdsplit(df, random_state = 888, split_size = [0.8, 0.1, 0.1]):
base_indices = np.arange(len(df))
base_indices = shuffle(base_indices, random_state = random_state)
nb_test = int(len(base_indices) * split_size[2])
nb_val = int(len(base_indices) * split_size[1])
test_idx = base_indices[0:nb_test]
valid_idx = base_indices[(nb_test):(nb_test+nb_val)]
train_idx = base_indices[(nb_test+nb_val):len(base_indices)]
print(len(train_idx), len(valid_idx), len(test_idx))
return train_idx, valid_idx, test_idx # split your data
train_idx, valid_idx, test_idx = Rdsplit(data.x, random_state = 888)
trainX = X[train_idx]
trainY = Y[train_idx]
validX = X[valid_idx]
validY = Y[valid_idx]
testX = X[test_idx]
testY = Y[test_idx]
# fit your model
clf = RegressionEstimator(n_outputs=trainY.shape[1],
fmap_shape1 = trainX.shape[1:],
dense_layers = [128, 64], gpuid = 0)
clf.fit(trainX, trainY, validX, validY)
# make prediction
testY_pred = clf.predict(testX)
rmse, r2 = clf._performance.evaluate(testX, testY)
print(rmse, r2)| Dataset | Task Metric | MoleculeNet (GCN Best Model) | Chemprop (D-MPNN model) | MolMapNet (MMNB model) |
|---|---|---|---|---|
| ESOL | RMSE | 0.580 (MPNN) | 0.555 | 0.575 |
| FreeSolv | RMSE | 1.150 (MPNN) | 1.075 | 1.155 |
| Lipop | RMSE | 0.655 (GC) | 0.555 | 0.625 |
| PDBbind-F | RMSE | 1.440 (GC) | 1.391 | 0.721 |
| PDBbind-C | RMSE | 1.920 (GC) | 2.173 | 0.931 |
| PDBbind-R | RMSE | 1.650 (GC) | 1.486 | 0.889 |
| BACE | ROC_AUC | 0.806 (Weave) | N.A. | 0.849 |
| HIV | ROC_AUC | 0.763 (GC) | 0.776 | 0.777 |
| PCBA | PRC_AUC | 0.136 (GC) | 0.335 | 0.276 |
| MUV | PRC_AUC | 0.109 (Weave) | 0.041 | 0.096 |
| ChEMBL | ROC_AUC | N.A. | 0.739 | 0.750 |
| Tox21 | ROC_AUC | 0.829 (GC) | 0.851 | 0.845 |
| SIDER | ROC_AUC | 0.638 (GC) | 0.676 | 0.68 |
| ClinTox | ROC_AUC | 0.832 (GC) | 0.864 | 0.888 |
| BBBP | ROC_AUC | 0.690 (Weave) | 0.738 | 0.739 |