Artificial intelligence based pathology as a biomarker of sensitivity to atezolizumab-bevacizumab in patients with hepatocellular carcinoma: a multicentric, retrospective study
Abstract
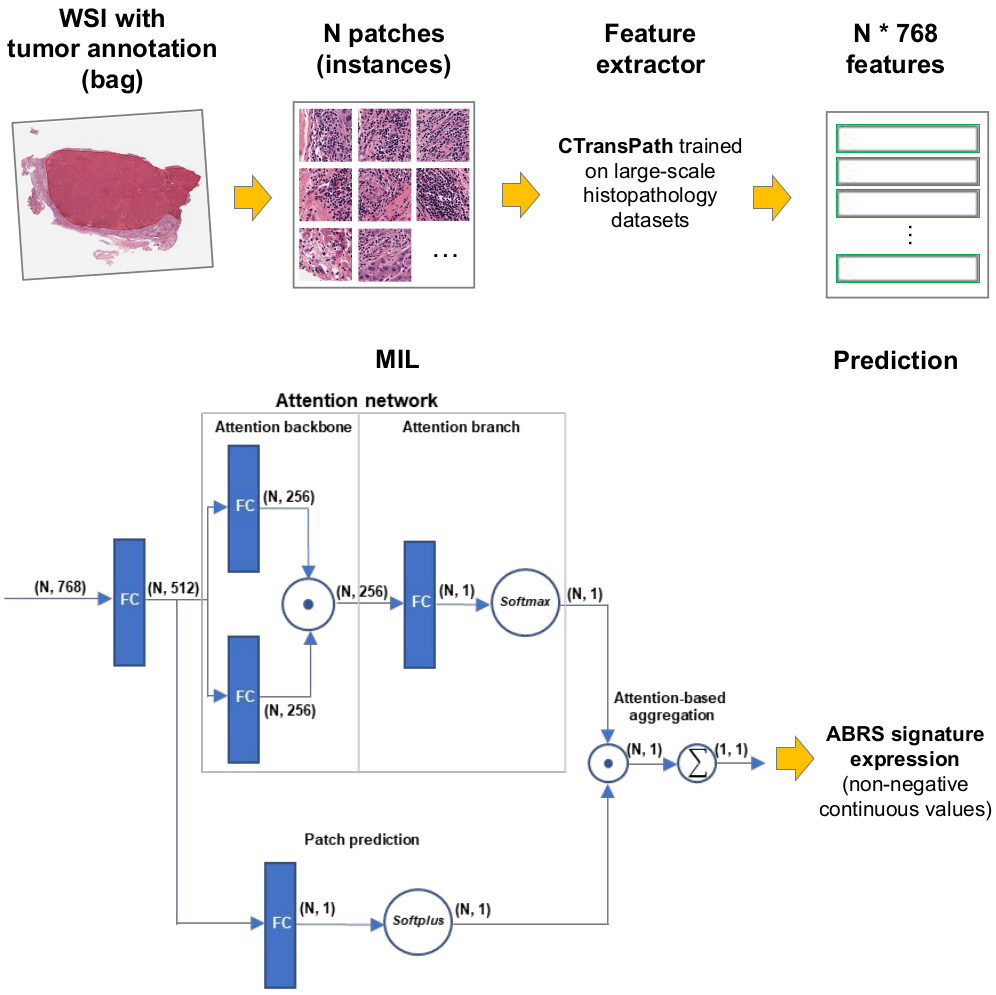
Biomarkers of sensitivity to targeted systemic therapies are key to improving the clinical management of patients. Gene signatures have been developed to this aim, however they remain very challenging to implement into clinical practice. Here, we develop a regression deep-learning model able to estimate, directly from digital histological slides, the expression of a gene signature linked to sensitivity to atezolimab-bevacizumab, the standard of care for advanced liver cancer. We further show that the model predicts progression-free survival in a multicentric series of treated patients. Integration of heat-maps obtained by deep learning and spatial transcriptomics finally confirms that the model recognizes areas with high expression of the signature and other immune related genes. This approach paves the way for the development of inexpensive biomarkers for targeted and immune therapies that could easily be implemented in clinical practice.
Installation
OS: Linux (Tested on Ubuntu 18.04)
Configure conda env and
Install the modified timm library
pip install timm-0.5.4.tarDownload CTransPath to the main directory as frozen feature extractor
More info
These codes are partially based on CLAM, CTransPath and Meylan et al. 2022.