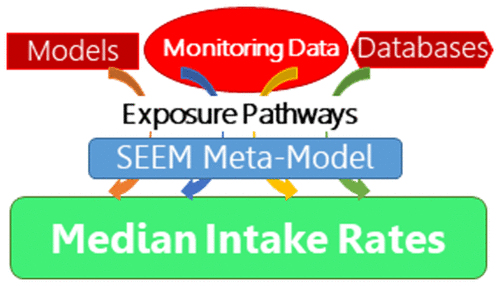
Systematic Empirical Evaluation of Models (SEEM)
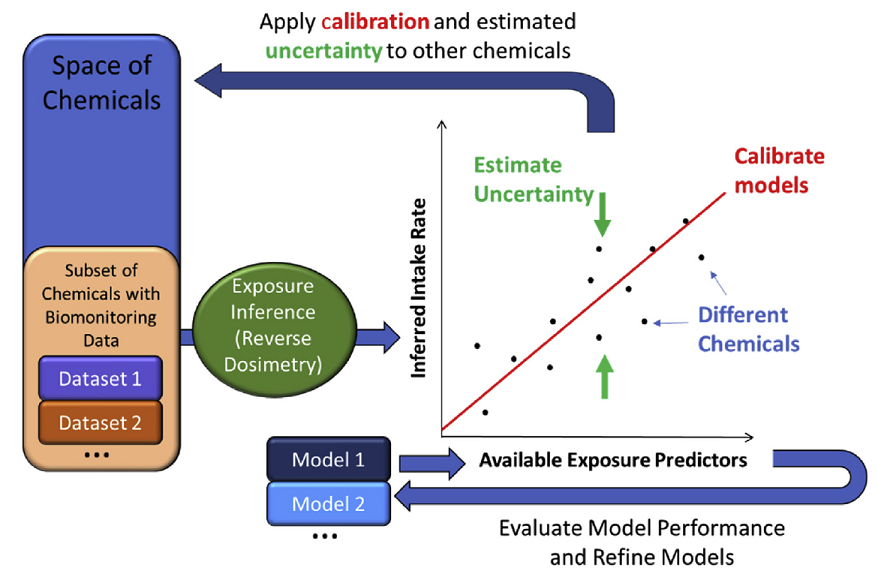
To apply high throughput exposure (HTE) models in a human health risk framework it is necessary to quantify the uncertainty in the HTE predictions. One recent uncertainty quantification approach has been to treat chemicals for which monitoring data are available as representative of chemicals without such data. In this way, the uncertainty of HTE predictions for chemicals without biomonitoring data may be estimated from chemicals with biomonitoring data. The predictions can be compared with population exposure biomonitoring data via toxicokinetic modeling, for purposes of evaluation and calibration. The ExpoCast project has made use of the Systematic Empirical Evaluation of Models (SEEM) framework, as illustrated in the figure (from: https://doi.org/10.1016/j.cotox.2019.07.001):
General Population Consensus Model (SEEM3)
For SEEM3 chemical structure and physicochemical properties were used to predict the probability that a chemical might be associated with any of four exposure pathways leading from sources–consumer (near-field), dietary, far-field industrial, and far-field pesticide–to the general population. The balanced accuracies of these source-based exposure pathway models range from 73 to 81%, with the error rate for identifying positive chemicals ranging from 17 to 36%. We then used exposure pathways to organize predictions from 13 different exposure models as well as other predictors of human intake rates. We created a consensus, meta-model using the SEEM framework in which the predictors of exposure were combined by pathway and weighted according to predictive ability for chemical intake rates inferred from human biomonitoring data for 114 chemicals. The consensus model yields an R2 of ∼0.8. This model was only calibrated for the total U.S. population. https://doi.org/10.1021/acs.est.8b04056
Demographic-Specific Heuristic Model (SEEM2)
For SEEM2 we developed a rapid heuristic method to determine potential human exposure to chemicals for application to the thousands of chemicals with little or no exposure data. We used Bayesian methodology to infer ranges of exposure consistent with biomarkers identified in urine samples from the U.S. population by the National Health and Nutrition Examination Survey (NHANES). We performed linear regression on inferred exposure for demographic subsets of NHANES demarked by age, gender, and weight using chemical descriptors and use information from multiple databases and structure-based calculators. Five descriptors are capable of explaining roughly 50% of the variability in geometric means across 106 NHANES chemicals for all the demographic groups, including children aged 6–11. https://doi.org/10.1021/es503583j
External Peer Review
In addition to being described in the peer-reviewed scientific literature an earlier version of these methods was reviewed by a Federal Insecticide, Fungicide, and Rodenticide Act (FIFRA) Scientific Advisory Panel (SAP) on "New High Throughput Methods to Estimate Chemical Exposure" in 2014: https://www.regulations.gov/docket?D=EPA-HQ-OPP-2014-0614
Authors
Principal Investigators
John Wambaugh [wambaugh.john@epa.gov] Caroline Ring [ring.caroline@epa.gov] Kristin Isaacs [isaacs.kristin@epa.gov] Katherine Phillips [phillips.katherine@epa.gov] Woodrow Setzer [setzer.woodrow@epa.gov]
SEEM3 Team
Jon Arnot Deborah Bennett Peter Egeghy Peter Fantke Lei Huang Olivier Jolliet Paul Price Hyeong-Moo Shin John Westgate https://doi.org/10.1021/acs.est.8b04056
SEEM2 Team
Anran Wang Kathie Dionisio Alicia Frame Peter Egeghy Richard Judson https://doi.org/10.1021/es503583j
License
License: GPL-3 https://www.gnu.org/licenses/gpl-3.0.en.html