This is a resource for software and information about alternative splicing. Contributions welcome...
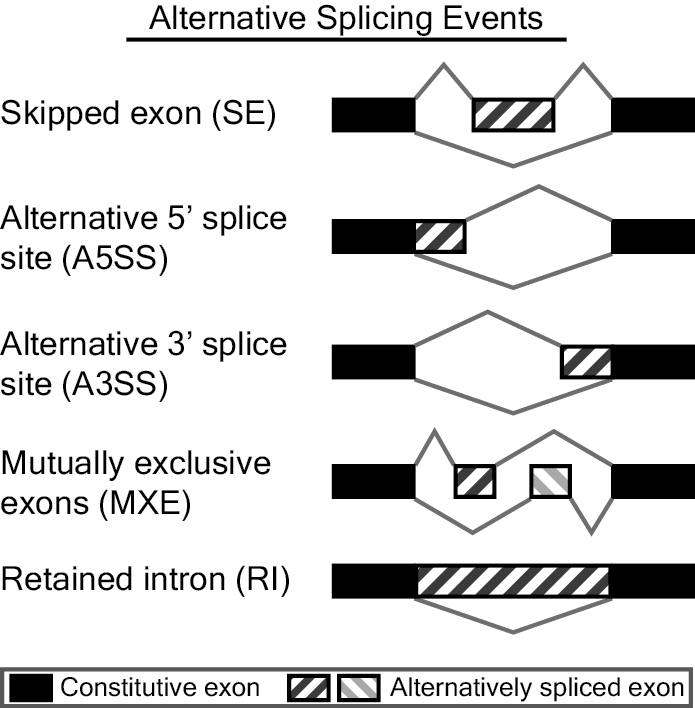
- Skipped exon or cassette exon (SE): An exon can be retained or spliced out of the primary transcript.
- Mutually exclusion exons (MXE): One of two exons is retained in mRNA splicing, but not both.
- Alternative 5' splice site (A5SS): An alternative 5' splice junction (donor site) is used that changes the 3' boundary of the upstream exon.
- Alternative 3' splice site (A3SS): An alternative 3' splice junction (acceptor site) is used that changes the 5' boundary of the downstream exon.
- Retained intron (RI): A sequence may be spliced out as an intron or retained, and there are no flanking introns.
Source: http://rnaseq-mats.sourceforge.net/rmats3.0.9/
- ALEXA-Seq - alternative expression analysis by massively parallel sequencing.
- AltAnalyze - analyze alternative splicing from single-cell and RNA-Seq data.
- Cufflinks - assemble and quantify transcripts.
- DEXSeq - identify differential exon usage.
- flotilla - reproduce machine learning analysis of gene expression and alternative splicing data.
- GMAP and GSNAP - detect complex variants and splicing in short reads, SNP-tolerant.
- G-Mo.R-Se - maps splice junctions to genome.
- HMMSplicer - discovery canonical and non-canonical splice junctions in short read datasets.
- JunctionSeq - identify differential splice junctions.
- MapSplice - map RNA-seq data to reference genome for splice junction discovery.
- MISO - determine alternative splicing expression.
- MMES - statistically determine alternative splicing.
- outrigger - calculate alternative splicing scores of RNA-Seq data based on junction reads and a de novo, custom annotation created with a graph database, especially made for single-cell analyses.
- rMATS - RNA-Seq Multavariate Analysis of Transcript Splicing. Reading rMATS output
- rmats2sashimiplot - visualize rMATS output using sashimi plots.
- SAW - identify splicing events from RNA-Seq data.
- Scripture - reconstruct transcript isoforms.
- SingleSplice - detect biological variation in alternative splicing within a population of single cells.
- SpliceMap - discover and align splice junctions for RNA-Seq reads.
- SpliceR - detect alternative splicing and predict coding potential.
- SplicingCompass - detect differential splicing using RNA-Seq data.
- SplitSeek - predict splice events from RNA-Seq data.
- STAR - identify alternative splicing.
- SUPPA - identify alternative splicing.
- TopHat - map splice junctions for RNA-Seq reads.
- ASIP - Alternative Splicing in Plants.
- ASG - Alternative Splicing Gallery for human genes.
- ASPicDB - Alternative Splicing PredICtion DataBase.
- ENSEMBL - Human and mouse genome annotations.
- FAST DB/Easana - Friendly Alternative Splicing and Transcripts Database.
- Hollywood exon annotation database - A website for querying a relational database of constitutive and alternative human exons, by using biological and descriptive features.
- HS3D - Data set of Homo Sapiens Exon, Intron and Splice regions extracted from GenBank Rel.123.
- H-DBAS - Human-transcriptome DataBase for Alternative Splicing.
- MAASE - Convenient access, identification, and annotation of alternative splicing events (ASEs), designed specifically with experimentalists in mind.
- Pro-Splicer - Alternative splicing database based on protein, mRNA, and EST Sequences.
- SpliceNest - Visualizing splicing of genes from EST Data for human, mouse, Drodophila and Arabidopsis.
| Tool | Performs split-read alignment | Transcript reconstruction (assembly) | Expression Analysis (any) | Gene expression analysis | Transcript specific expression analysis | Exon junction expression | Quantitative alternative expression analysis | Expression level sensitivity | Output | Minimum read length required or recommended | Visualization tool | Performs comparisons between conditions (ex. tumor vs normal) | Relevant comparison to Alexa-seq | Data type supported | Citation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alexa-Seq | N | N | Y | Y | Y | Y | Y | Junction | "Expression and structure information for junctions and genes; UCSC track info | extensive alternative expression visualization / statistics / graphs" | No minimum (tested on 36bp-100bp reads) | "Extensive | includes custom graphs and links to UCSC browser" | Y | - |
| Cufflinks | N | Y | Y | Y | Y | N | Y | Transcript | "Transcript information and expression statistics | BED | GTF" | 75bp | UCSC browser | Y | Y |
| Scripture | N | Y | Y | N | N* | N | N | Transcript* | "Transcript structure information and non-parsimonious expression statistics | BED" | 75bp | UCSC browser | N | N | Illumina |
| SpliceMap | N | N | Y | Y | N | Y | N | Junction | "Alignments. SAM | BED | Wig" | 50bp | UCSC browser | N | N |
| TopHat | Y | N | N | N | N | Y | N | Junction | "Alignments. SAM | BED | Wig" | 75bp | N | N | N |
| MMES | Y | N | N | N | N | Y | N | Junction | Identified splice junctions and p-values | 25bp | N | N | N | Illumina | "Wang et al. |
| G-Mo.R-Se | Y | Y | N | N | N | N | N | Transcript* | Transcript structure information. GFF | 25bp | Grape Genome Browser | N | N | Illumina | "Denoeud et al. |
| SplitSeek | Y | N | N | N | N | Y | N | Junction | Alignments. BED | 50bp | UCSC browser | N | N | SOLiD only | "Ameur et al.2010 |
| GSNAP | Y | Y | N | N | N | N | N | N/A | Alignments. SAM and FASTA | Minimum 14bp (tested on 36bp reads) | UCSC browser | N | N | Illumina | sodium bisulfite-treated DNA sequencing (for analysis of methylation status) |
| Tool | Performs split-read alignment | Transcript reconstruction (assembly) | Expression Analysis (any) | Gene expression analysis | Transcript specific expression analysis | Exon or Junction expression | Quantitative alternative expression analysis | Expression level sensitivity | Output | Minimum read length required or recommended | Ability to identify rearrangements / indels | Junction Identification | Implementation | Public tool | Open Source |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alexa-seq | N | N | Y | Y | Y | Y | Y | Junction | "Expression and structure information for junctions and genes; UCSC track info | extensive alternative expression visualization / statistics / graphs" | No minimum (tested on 36bp-100bp reads) | N | Database | Perl/R/Unix | Y |
| Cufflinks | N | Y | Y | Y | Y | N | Y | Transcript | "Transcript information and expression statistics | BED | GTF" | 75bp | N | Predicted from data | C++ |
| Scripture | N | Y | Y | N | N* | N | N | Transcript* | "Transcript structure information and non-parsimonious expression statistics | BED" | 75bp | N | Predicted from data | Java | Y |
| SpliceMap | N | N | Y | Y | N | Y | N | Junction | "Alignments. SAM | BED | Wig" | 50bp | N | Predicted from data | Python |
| TopHat | Y | N | N | N | N | Y | N | Junction | "Alignments. SAM | BED | Wig" | 75bp | N | Predicted from data | C++/Python |
| MMES | Y | N | N | N | N | Y | N | Junction | Identified splice junctions and p-values | 25bp | N | Predicted from data | Published algorithm only | N | N/A |
| G-Mo.R-Se | Y | Y | N | N | N | N | N | Transcript* | Transcript structure information. GFF | 25bp | N | Predicted from data | Perl | Y** | Y |
| SplitSeek | Y | N | N | N | N | Y | N | Junction | Alignments. BED | 50bp | Y | Predicted from data | Perl | Y | Y (GPL) |
| GSNAP | Y | Y | N | N | N | N | N | N/A | Alignments. SAM and FASTA | Minimum 14bp (tested on 36bp reads) | Y | Database or predicted form data | "Source code in C | utility programs in Perl" | Y |
| Tool | Performs split-read alignment | Transcript reconstruction (assembly) | Expression Analysis (any) | Gene expression analysis | Transcript specific expression analysis | Exon or Junction expression | Quantitative alternative expression analysis | Expression level sensitivity | Output | Minimum read length required or recommended | Ability to identify rearrangements / indels | Junction Identification | Implementation | Public tool | Open Source |
| Alexa-seq | N | N | Y | Y | Y | Y | Y | Junction | "Expression and structure information for junctions and genes; UCSC track info | extensive alternative expression visualization / statistics / graphs" | No minimum (tested on 36bp-100bp reads) | N | Database | Perl/R/Unix | Y |
| Cufflinks | N | Y | Y | Y | Y | N | Y | Transcript | "Transcript information and expression statistics | BED | GTF" | 75bp | N | Predicted from data | C++ |
| Scripture | N | Y | Y | N | N* | N | N | Transcript* | "Transcript structure information and non-parsimonious expression statistics | BED" | 75bp | N | Predicted from data | Java | Y |
| SpliceMap | N | N | Y | Y | N | Y | N | Junction | "Alignments. SAM | BED | Wig" | 50bp | N | Predicted from data | Python |
| TopHat | Y | N | N | N | N | Y | N | Junction | "Alignments. SAM | BED | Wig" | 75bp | N | Predicted from data | C++/Python |
| MMES | Y | N | N | N | N | Y | N | Junction | Identified splice junctions and p-values | 25bp | N | Predicted from data | Published algorithm only | N | N/A |
| G-Mo.R-Se | Y | Y | N | N | N | N | N | Transcript* | Transcript structure information. GFF | 25bp | N | Predicted from data | Perl | Y** | Y |
| SplitSeek | Y | N | N | N | N | Y | N | Junction | Alignments. BED | 50bp | Y | Predicted from data | Perl | Y | Y (GPL) |
| GSNAP | Y | Y | N | N | N | N | N | N/A | Alignments. SAM and FASTA | Minimum 14bp (tested on 36bp reads) | Y | Database or predicted form data | "Source code in C | utility programs in Perl" | Y |
* Transcript expression values are non-parsimonious
** Not supported (current version not stable)