Table of Content:
Parallel Approximate Bayesian Computing Sequential Monte Carlo (pABC-SMC) algorithm for statistical inference for multi-scale and multi-cellular biological processes
The pABC-SMC repository provides an implementation for the simulation of tumour spheroids. Furthermore, it facilitates the statistical inference of model parameters from spheroid growth curves and histological information. Its key features are
efficient numerical implementation for lattice-based model for tumour spheroid growth; collection of experimental data for SK-MES-1 cells; statistical inference using Approximate Bayesian Computing Sequential Monte Carlo (ABC-SMC); and parallelisation using the ABC-SMC algorithm for our grid topology.
Algorithms implemented in the pABC-SMC repository employ C++ and MATLAB. To exploit its functionality the MATLAB Statistical Toolbox. The parallel implementation is tailored for our computing grid. The use of other infrastructures requires reimplementation.
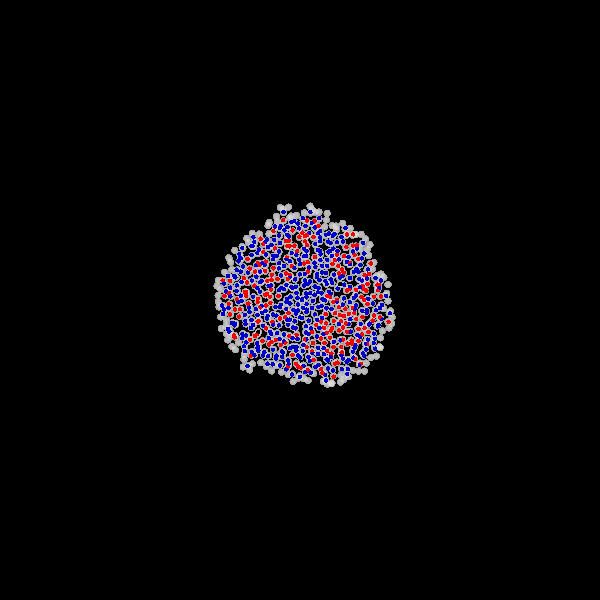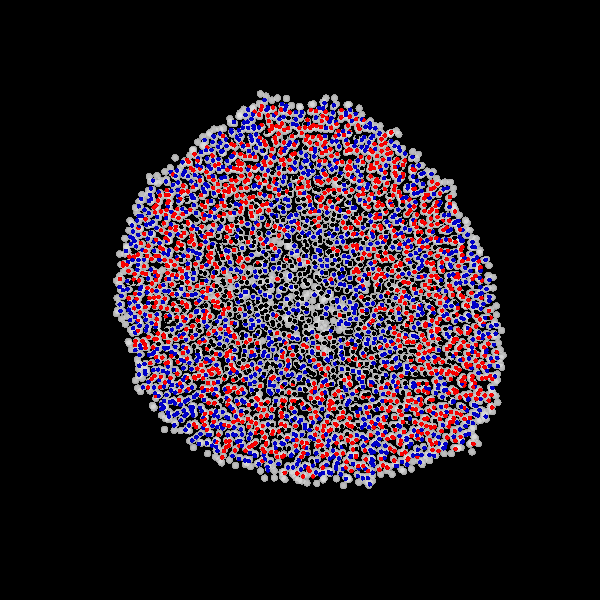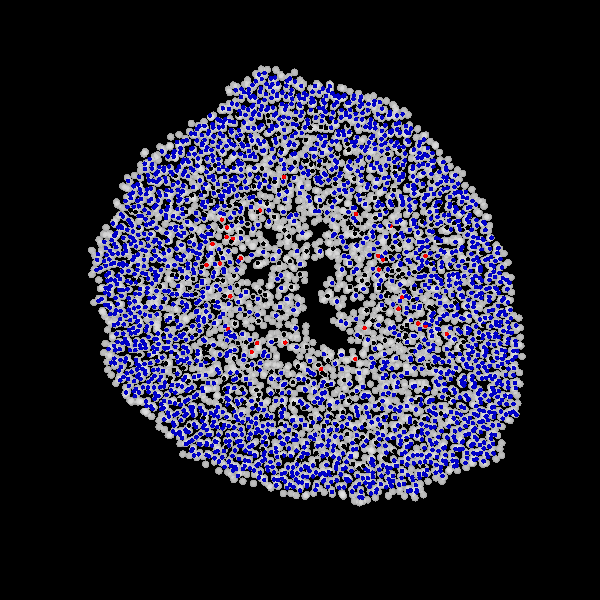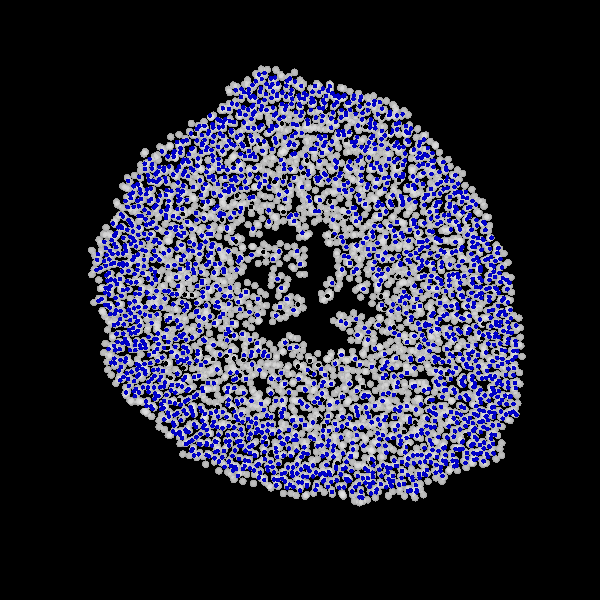
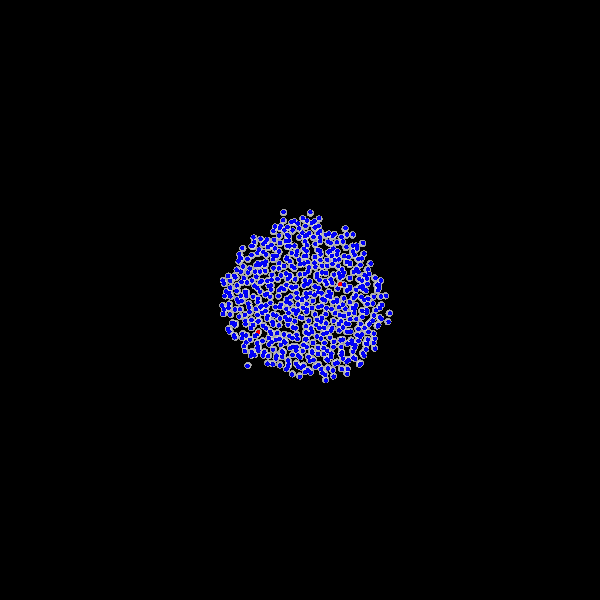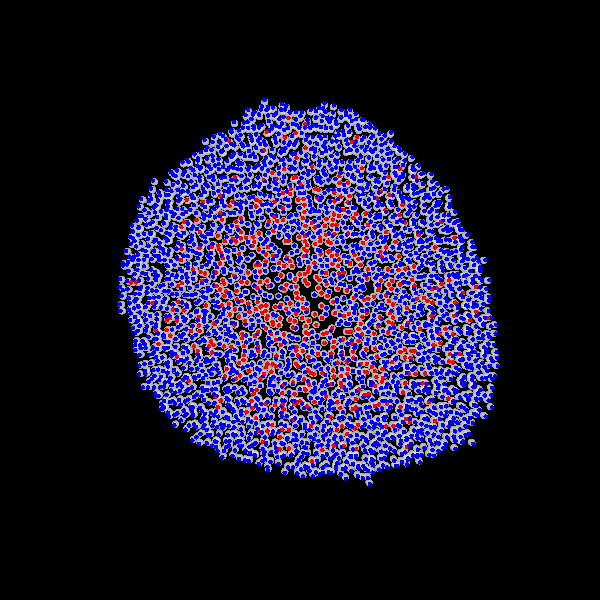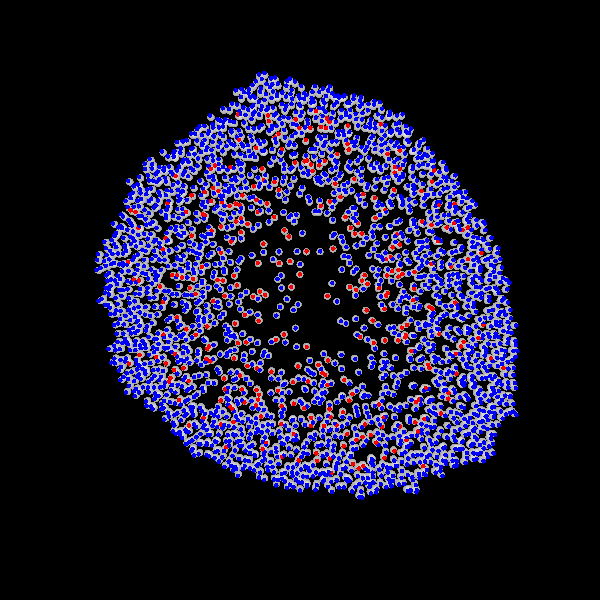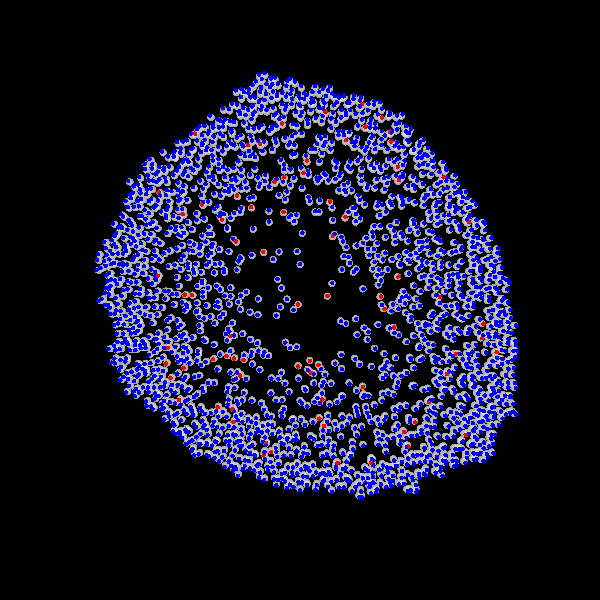
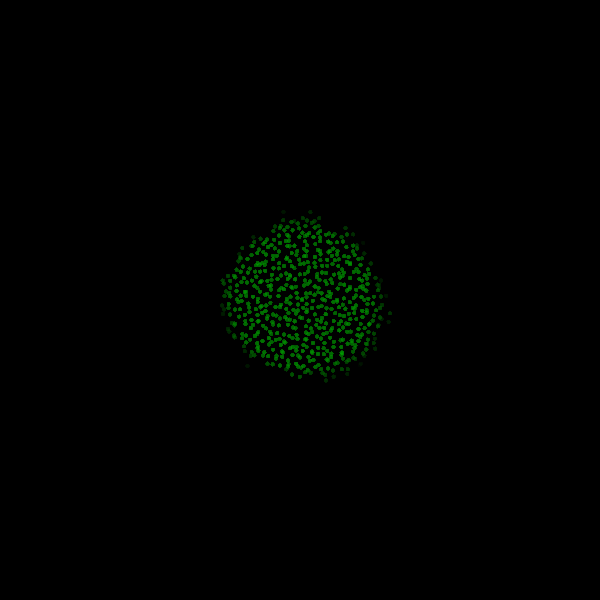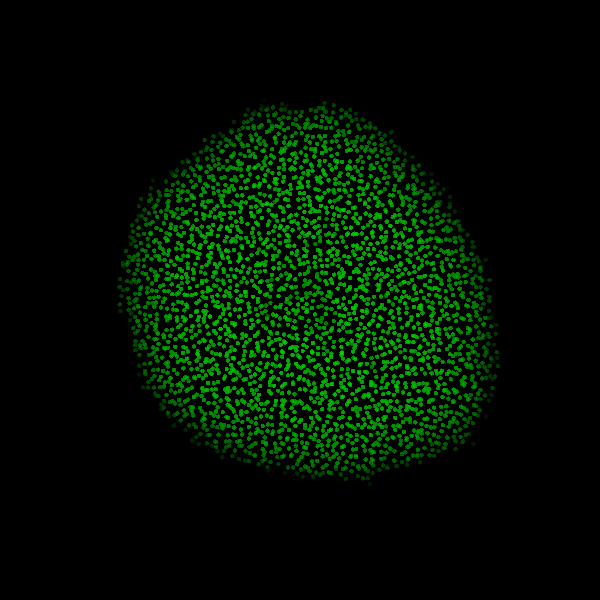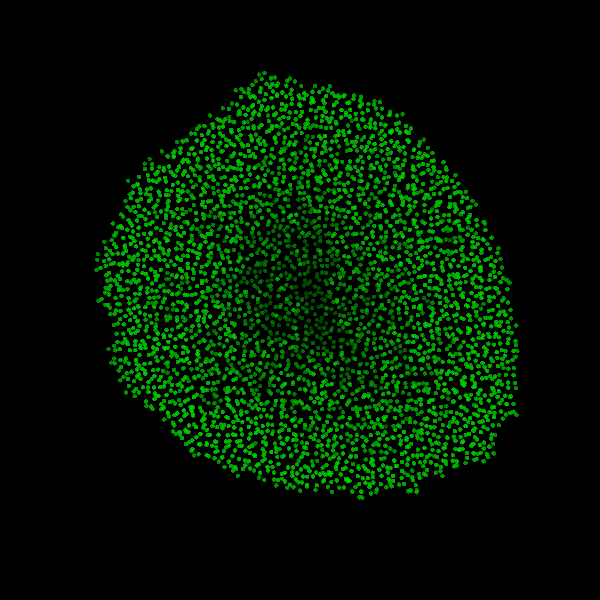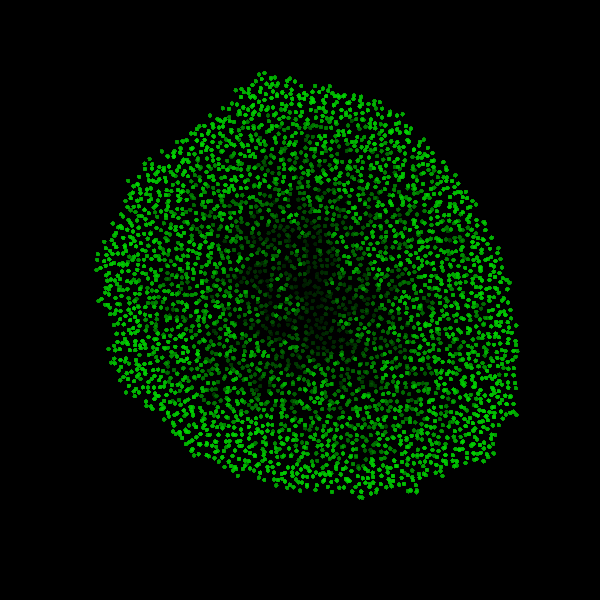
Fig. Figure is showing an example simulation of a tumor spheroid growing for 40 days. *left:* dividing cells (red=dividing, blue=quiescent). *left:* necrotic cells (red=necrotic, blue=alive). *left:* extracellular matrix (ECM).No prerequisites are needed for compilation, except autotools, make, g++ and gsl (the gnu scientific library).
To configure and compile the code on your mac system execute:
automake --add-missing
autoreconf
./configure
make
sudo make install
Please note that the OSX version El Capitan broke linking against the GSL; the makefile provided will not suffice in this case & compilation can not be supported.
On Linux, only
./configure
sudo make install
are required.
If everything worked properly, the following files will have been installed:
-
Library: libnix
-
Executables:
- nix-tumor2d
- nix-tumor3d
- nix-compare2d
- nix-compare3d
####1) Tumor Simulation: For a simple tumour growth simulation of 1 realisation for 200 hours on a two-dimensional lattice execute:
nix-tumor2d -x 1 -y 200
The equivalent in three dimensions and a batch of 3 realisations:
nix-tumor3d -x 3 -y 200
The two-dimensional simulation will take a few minutes while the three dimensional implementation several simulation will take several days.
The default model parameter values can be modified by passing further arguments:
######Table 1: Model Parameters
| Argument | Description | Default Value |
|---|---|---|
-Rdiv[FLOAT] |
division rate (1/hours) | 0.032 |
-RReentranceProbabilityLength[FLOAT] |
division depth (\mu m) | 130 |
-RInitialRadius[FLOAT] |
initial tumor radius (\mu m) | 1 |
-RInitialQuiescentFraction[FLOAT] |
initial quiescent cell fraction (-) | 0.032 |
-RECMProductionRate[FLOAT] |
ECM production rate (1/hours) | 0.032 |
-RECMDegradationRate[FLOAT] |
ECM degradation rate (1/hours) | 0.032 |
-RECMThresholdQuiescence[FLOAT] |
ECM division threshold (-) | 0.032 |
-Rre[FLOAT] |
cell cycle reentrance rate (1/hours) | 0.032 |
-Rnec[FLOAT] |
necrosis rate (1/hours) | 0.032 |
-Rlys[FLOAT] |
lysis rate (1/hours) | 0.032 |
-RATPThresholdQuiescence[FLOAT] |
ATP synthesis division threshold (mM/h) | 0.032 |
-RATPThresholdDeath[FLOAT] |
ATP synthesis necrosis threshold (mM/h) | 0.032 |
-RLactateThresholdQuiescence[FLOAT] |
lactate division threshold (mM) | 0.032 |
-RLactateThresholdDeath[FLOAT] |
lactate necrosis threshold (mM) | 0.032 |
-RWasteDiffusion[FLOAT] |
waste diffusion coefficient (\mu m^2/hours) | 0.032 |
-RWasteUptake[FLOAT] |
waste degradation rate (1/hours) | 0.032 |
-RWasteThresholdSlowedGrowth[FLOAT] |
waste division threshold (mM) | 0.032 |
-RWasteIntoxicatedCellCycles[FLOAT] |
max #cell cycles under waste exposure / O2 deprivation | 0.032 |
For a complete list of program arguments run:
nix-tumor3d -h
####2) Comparison with Data: In order to compare the simulation results on the fly with given data, the following command can be used instead:
nix-compare2d DATA_FILES LIKELIHOOD_THRESHOLD PARAMETER_LIST
-
The
DATA_FILESare passed asArgument Description -g[FILENAME]growth curve** -k[FILENAME]day 17: KI67 positive / proliferating cell fraction* -t[FILENAME]day 17: TUNEL positive / necrotic cell fraction* -e[FILENAME]day 17: COLIV intensity / extra-cellular matrix (ECM) density* -K[FILENAME]day 24: KI67 positive / proliferating cell fraction* -T[FILENAME]day 24: TUNEL positive / necrotic cell fraction* -E[FILENAME]day 24: COLIV intensity / extra-cellular matrix (ECM) density* * as function of the distance to the outer tumuor border (\mu m) ** as function of time (days) -
The
LIKELIHOOD_THRESHOLDindicates the value which will stop a running simulation if exceeded and will be returned back. It is passed as-l[FLOAT] -
PARAMETER_LISTis an optional list of 0 to 18 values corresponding to the model parameters in Tab. 1FLOAT ... FLOAT
After finishing the simulation for the given model parameters the program will print the likelihood to stdout.
The directory \data contains experimental data for four experimental conditions.
The measured quantities are:
- The spheroid radius (GC) as a function of time.
- The fraction of proliferating cells on day 17 (T3_Ki67) and day 24 (T4_Ki67) as a function of the distance from the spheroid rim.
- The fraction of necrotic cells on day 17 (T3_TUNEL) and day 24 (T4_TUNEL) as a function of the distance from the spheroid rim.
- The extracellular matrix intensity on day 17 (T3_ECM) and day 24 (T4_ECM) as a function of the distance from the spheroid rim.
These quantities are reported for up to four experimental conditions conditions:
- Condition I: glucose concentration = 1 mM, oxygen concentration = 0.28 mM
- Condition II: glucose concentration = 25 mM, oxygen concentration = 0.28 mM
- Condition III: glucose concentration = 5 mM, oxygen concentration = 0.28 mM
- Condition IV: glucose concentration = 25 mM, oxygen concentration = 0.07 mM
As the number of replicates available for the histological data is rather same and the estimated standard deviation therefore not very reliable. For parameter estimation, we considered therefore in addition to standardalternative distance measures.
The experimental data are stored in the files \data\*.dat. The files \data\*.dat.* provide alternative measures for uncertainty in the third column. The max-min distance and the standard deviation provided in *.dat.MinMax and *.dat.Std are calculated over all time points / distances for a given dataset.
| Files | 1st column | 2nd column | 3rd column |
|---|---|---|---|
SK-MES1_*.dat |
time (h) / distance (\mu m) | mean | standard deviation |
SK-MES1_*.dat.Mean |
time (h) / distance (\mu m) | mean | mean / 10 |
SK-MES1_*.dat.Std |
time (h) / distance (\mu m) | mean | standard deviation of mean |
SK-MES1_*.dat.MinMax |
time (h) / distance (\mu m) | mean | max(mean) - min(mean) |
** An example: ** Comparison of the simulation result of the 2D model for an oxygen concentration of 0.28 mM and a glucose concentration of 25 mM to the corresponding dataset (III) using the dynamic range of the measurement (max - min) for weighting
nix-compare2d -O0.28 -G25 \
-gdata/SK-MES1_III_GC.dat.MinMax \
-kdata/SK-MES1_III_T3_Ki67.dat.MinMax -Kdata/SK-MES1_III_T4_Ki67.dat.MinMax \
-tdata/SK-MES1_III_T3_TUNEL.dat.MinMax -Tdata/SK-MES1_III_T4_TUNEL.dat.MinMax \
-edata/SK-MES1_III_T3_ECM.dat.MinMax -Edata/SK-MES1_III_T4_ECM.dat.MinMax
The current implementation of the pABC-SMC algorithm is very problem and infrastructure specific. We will are currently working on a more flexible implementation of the code. We are however happy to provide the current implementation upon request.
- Nick Jagiella - nick.jagiella@gmail.com (main developer)
- Dennis Rickert - dennis.rickert@helmholtz-muenchen.de (developer)
- Jan Hasenauer - jan.hasenauer@helmholtz-muenchen.de
If you use this software in a publication, please cite one of the following manuscript:
- N. Jagiella, B. Müller, M. Müller, I. E. Vignon-Clementel and D. Drasdo. Inferring growth control mechanisms in growing multi-cellular spheroids of NSCLC cells from spatial-temporal image data, PLoS Comput. Biol., 12(2): e1004412 , 2016.