This is the code of the Non-peptidic epitope prediction tool (NP_epitope_predictor). A running version of the tool is available at: http://tools-staging.iedb.org/np_epitope_predictor/ Licensed under the Non-Profit Open Software License version 3.0.
git clone https://github.com/paulzierep/NP_epitope_predictor.git
Install conda:
Activate a new environment:
conda activate **env_name**
Install all packages:
conda install --file requirements.txt
Unfortunately rdkit is difficult to set up outside the coda env. Therefore this is not tested. In general it should work to install rdkit with any method described here:
Then install the other packages with
pip install -r requirements.txt
In order to run a prediction it is not necessary to fit the predictor, it can run with pre-fitted data, which is provided in:
./tools/ML_data.zip
Unzip in same folder:
cd ./tools/
unzip ML_data.zip
An example how to run the prediction is given in the script:
./tools/run_prediction.py
The predictor needs as input a correct SMILES string e.g.: "CCCCCCO".
There are two optional arguments which are currently included:
| Argument | Parameter | Explanation |
|---|---|---|
| only_epitopes | True | The most similar epitopes are listed, |
| only_epitopes | False | The most similar compounds from the entire ChEBI are listed |
| sort_order | "E" | Uses the eucleadien distance as similarity measure for the target compounds meaning the shown compounds have the most FPs in common with the query molecule |
| sort_order | "T" | Uses the overall Tanimoto Coefficient as similarity measure, meaning the shown compound are most similar to the query molecule, this requires, that the similarity is computed between all compounds in the cluster and the query (takes a bit longer) |
The predictor returns a dict which holds all needed information, e.g.: Class, B_cell probability...
A tree of the dict is shown below. The ontology and fp_imp (FP importance) are pandas data frames. They can be converted into html, dict or json with (.to_html(),.to_dict() or to_json()).
error <class 'NoneType'>
input_svg <class 'str'>
cluster_info <class 'dict'>
Cluster <class 'int'>
Members <class 'int'>
Warning <class 'float'>
Name <class 'str'>
Ontology <class 'pandas.core.frame.DataFrame'>
classify_info_b <class 'dict'>
proba <class 'float'>
fp_imp <class 'pandas.core.frame.DataFrame'>
fitting_info <class 'NoneType'>
p_support <class 'int'>
n_support <class 'int'>
classify_info_t <class 'dict'>
proba <class 'float'>
fp_imp <class 'pandas.core.frame.DataFrame'>
fitting_info <class 'NoneType'>
p_support <class 'int'>
n_support <class 'int'>
The utility functions: Results_To_Json creates a JSON object from the results dict. The json tree can be observed for example by copy-and-pasting into http://jsonviewer.stack.hu/
The utility functions: Results_To_Html creates one possibility of how the HTML output could look like. Although the incorporation of the json into a framework would be preferable.
If the SMILES can not be parsed, the parsing error is recorded and stored in the result dict in "error". This can be shown to the user, as it is currently implemented in Results_To_Json and Results_To_Html.
To simplify the useage of the NP_epitope_predictor a lightweight django wrapper is also available. The wrapper allows for the handling of input and output without the need to use the python API. A running version of the wrapper is currently hosted at: http://tools-staging.iedb.org/np_epitope_predictor/
The wrapper can be used via:
conda activate **env_name**
cd tool/django_wrapper/np_epitope_predictor
python manage runserver
Open the wrapper in a web browser via http://127.0.0.1:8000/np_epitope_predictor/
The update requires to download 3 files:
A CSV for each epitope type, i.e. B cells and T cells from the IEDB and the SDF file of the ChEBI 3 star dataset (ChEBI_complete_3star.sdf).
The update data is stored in the folder NP_epitope_predictor_gitlab/tool/ML_data_updated; this folder is not shared via git (too large), so the ML_data_updated.zip file needs to be unzipped first.
The folder ML_data should not be touched, keep the original data for the publication.
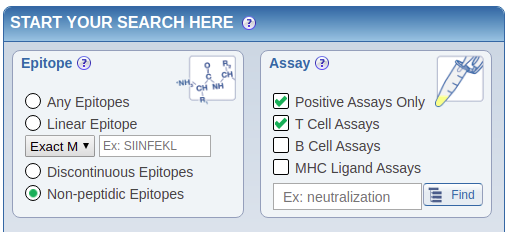
Download the epitopes via an IEDB query:
- Go to https://www.iedb.org/
 |
|---|
| Select the epitopes |
 |
|---|
 |
| :--: |
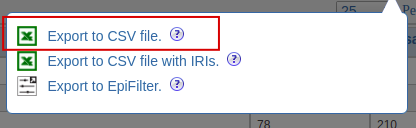
| Export csv |
-
Copy the generated file (e.g. epitope_table_export_1590139338.zip) into the update folder (NP_epitope_predictor_gitlab/tool/ML_data_updated/epitope_update_input/**update_date**)
-
Unzip the file and rename into epitope_table_b_cell_pos.csv
-
Repeat the same process for T cells
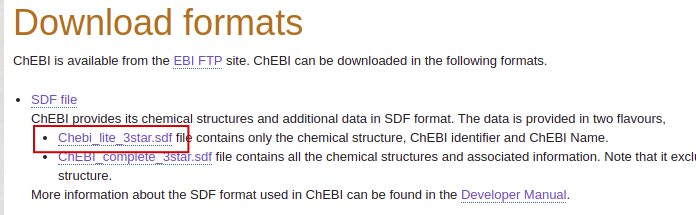
Download the CSV file from ChEBI:
 |
|---|
| Downoload the SDF file |
- Unzip and copy into the update folder (NP_epitope_predictor_gitlab/tool/ML_data_updated/epitope_update_input/**update_date**)
-
The update folder (NP_epitope_predictor_gitlab/tool/ML_data_updated/epitope_update_input/**update_date**) should contain the files:
- epitope_table_b_cell_pos.csv
- epitope_table_t_cell_pos.csv
- ChEBI_lite_3star.sdf
-
Set the path of the update folder in the update script (update_predictor.py) (NP_epitope_predictor_gitlab/tool/ML_data_updated/epitope_update_input/**update_date**)
#this is where the input data (IEDB csv and ChEBI sdf) is located UPDATE_PATH = os.path.join(DATA_PATH, 'epitope_update_input', '05-11-2020') -
Run the update script (update_predictor.py), approx.: 1780.9s
-
This should generate all the new files in ML_data_updated. The django wrapper uses this files. No further modifications needed. The online view will now compute results based on the new data.
sudo docker build -t NP_epitope_predictor:1.0 . --no-cache
- Push docker image to gitlab