MatchingFrontier: Computation of the Balance-Sample Size Frontier in Matching Methods for Causal Inference
MatchingFrontier implements the methods described in King, Lucas, and
Nielsen (2017) for exploring the balance-sample size frontier in
observational studies, a function the relates the size of a matched
subset to the covariate balance attained. Included are function to
create the frontier, visualize it, and estimate treatment effects along
it. In addition, functions are included to export a matched dataset at a
single point on the frontier and assess model dependence for a single
treatment effect estimate. MatchingFrontier interfaces with MatchIt
to provide additional tools for assessing balance in matched datasets
and estimating effects after matching.
Below is an example of using the MatchingFrontier to examine the
balance-sample size frontier for the effect of a job training program on
earnings. See vignette("MatchingFrontier") for more information on the
setup and a more detailed exposition.
library("MatchingFrontier")
data("lalonde", package = "MatchIt")
mahal.frontier <- makeFrontier(treat ~ age + educ + race + married +
nodegree + re74 + re75,
data = lalonde,
QOI = "FSATT",
metric = "dist",
verbose = FALSE)
mahal.frontier#> A matchFrontier object
#> - quantity of interest: FSATT
#> - imbalance metric: average pairwise distance
#> - treatment: treat
#> - covariates: age, educ, race, married, nodegree, re74, re75
#> - number of points: 154
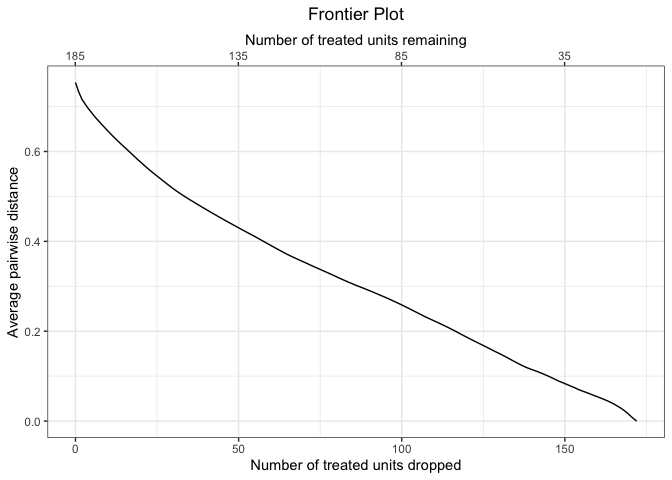
Plotting the frontier provides a clear picture of the relationship between the number of units pruned in the matching and the remaining imbalance as measured by the average Mahalanobis imbalance.
plot(mahal.frontier)We can then estimate effects along the frontier.
mahal.estimates <- estimateEffects(mahal.frontier,
base.form = re78 ~ treat,
verbose = FALSE)
mahal.estimates#> A frontierEstimates object
#> - quantity of interest: FSATT
#> - model sensitivity method: none
#> - number of estimates: 149
#> - treatment: treat
#> - covariates: age, educ, race, married, nodegree, re74, re75
#> - outcome model: re78 ~ treat
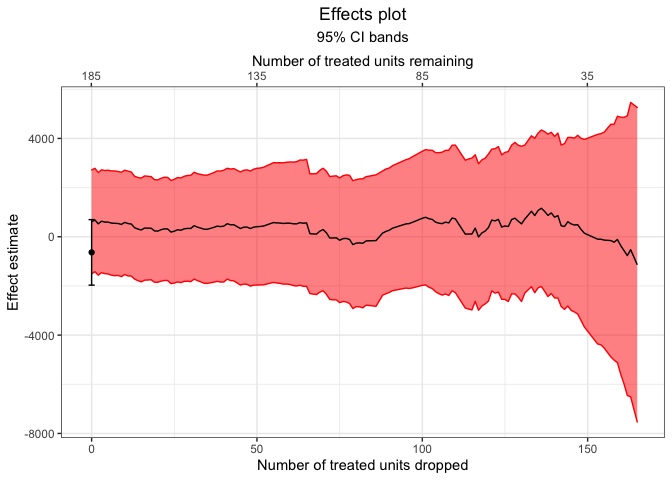
Finally, we can plot the estimates and their confidence intervals.
plot(mahal.estimates)Other tools include generateDataset() and frontier_to_matchit() for
extracting a matched dataset at one point on the frontier and
modelDependence() for computing model dependence bounds for a single
estimate. See vignette("MatchingFrontier") for a more in-depth
tutorial.
The work in this package is based off the following paper:
King, Gary, Christopher Lucas, and Richard A. Nielsen. 2017. “The Balance-Sample Size Frontier in Matching Methods for Causal Inference.” American Journal of Political Science 61(2): 473–89.