(work in progress)
AlphaFold
This repository contains a minimal example of DeepMind's AlphaFold 2 model, that runs the model with no data and third-party requirements, for a particular protein sequence 6y4f. This repository uses their code published at deepmind/alphafold.
This code runs on CPU by assumption, but it can also be run using a GPU (tested using a tensorflow-gpu docker image with a cuda-compatible version of jax installed).
The input features from a model run using a fasta sequence from the link above on a non-gpu based google cloud instance are provided in the 6y4f folder.
Setup
conda create -n alphafold python=3.7 && conda activate alphafold
conda install -y -c conda-forge openmm=7.5.1 pdbfixer
pip install -r requirements.txt
cd ~/miniconda3/envs/alphafold/lib/python3.7/site-packages
patch -p0 < ~/alphafold/openmm.patch && cd -
# pip install --upgrade jax jaxlib==0.1.69+cuda111 -f https://storage.googleapis.com/jax-releases/jax_releases.html
# download model params belowModel parameters
The AlphaFold parameters are available from https://storage.googleapis.com/alphafold/alphafold_params_2021-07-14.tar. The following models will be downloaded:
- 5 models which were used during CASP14, and were extensively validated for structure prediction quality (see Jumper et al. 2021, Suppl. Methods 1.12 for details).
- 5 pTM models, which were fine-tuned to produce pTM (predicted TM-score) and predicted aligned error values alongside their structure predictions (see Jumper et al. 2021, Suppl. Methods 1.9.7 for details).
ROOT_DIR="data/params"
SOURCE_URL="https://storage.googleapis.com/alphafold/alphafold_params_2021-07-14.tar"
BASENAME=$(basename "${SOURCE_URL}")
wget -P "${ROOT_DIR}" "${SOURCE_URL}"
tar -xvpf "${ROOT_DIR}/${BASENAME}" -C "${ROOT_DIR}"
rm "${ROOT_DIR}/${BASENAME}"
Running AlphaFold
Running replication_script.py should output the following expected outputs.
AlphaFold output
The outputs of the script above include the computed MSAs
unrelaxed structures, relaxed structures, raw model outputs.
The output directory will have the following structure:
output_dir/
ranked_0.pdb
The contents of each output file are as follows:
ranked_*.pdb– A PDB format text file containing the relaxed predicted structures, after reordering by model confidence. Hereranked_0.pdbshould contain the prediction with the highest confidence. To rank model confidence, DeepMind uses predicted LDDT (pLDDT), see Jumper et al. 2021, Suppl. Methods 1.9.6 for details.
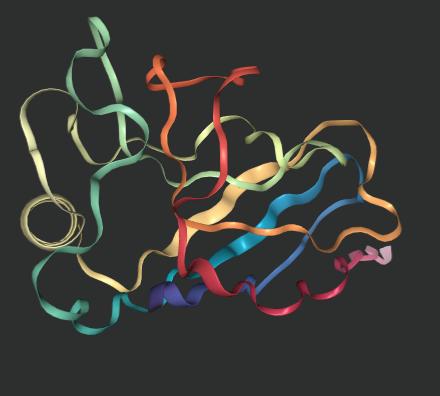
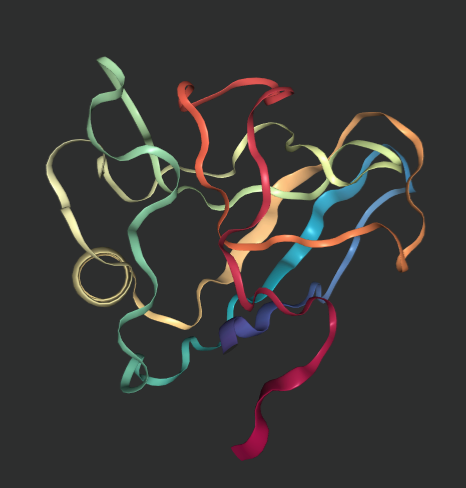
The replicated outputs of this repository are shown below (nglviewer outputs images that are displayed in jupyter notebooks).
from Bio.PDB import PDBParser
from nglview import show_biopython
file_loc = '...'
def get_view(file):
return show_biopython(PDBParser().get_structure('6y4f', file_loc + file))
get_view('6y4f/6y4f.pdb') # true
get_view('6y4f/ranked_0.pdb') # predictedLicense and Disclaimer
This is not an officially supported Google product.
Copyright 2021 DeepMind Technologies Limited.
AlphaFold Code License
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at https://www.apache.org/licenses/LICENSE-2.0.
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
Model Parameters License
The AlphaFold parameters are made available for non-commercial use only, under the terms of the Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0) license. You can find details at: https://creativecommons.org/licenses/by-nc/4.0/legalcode