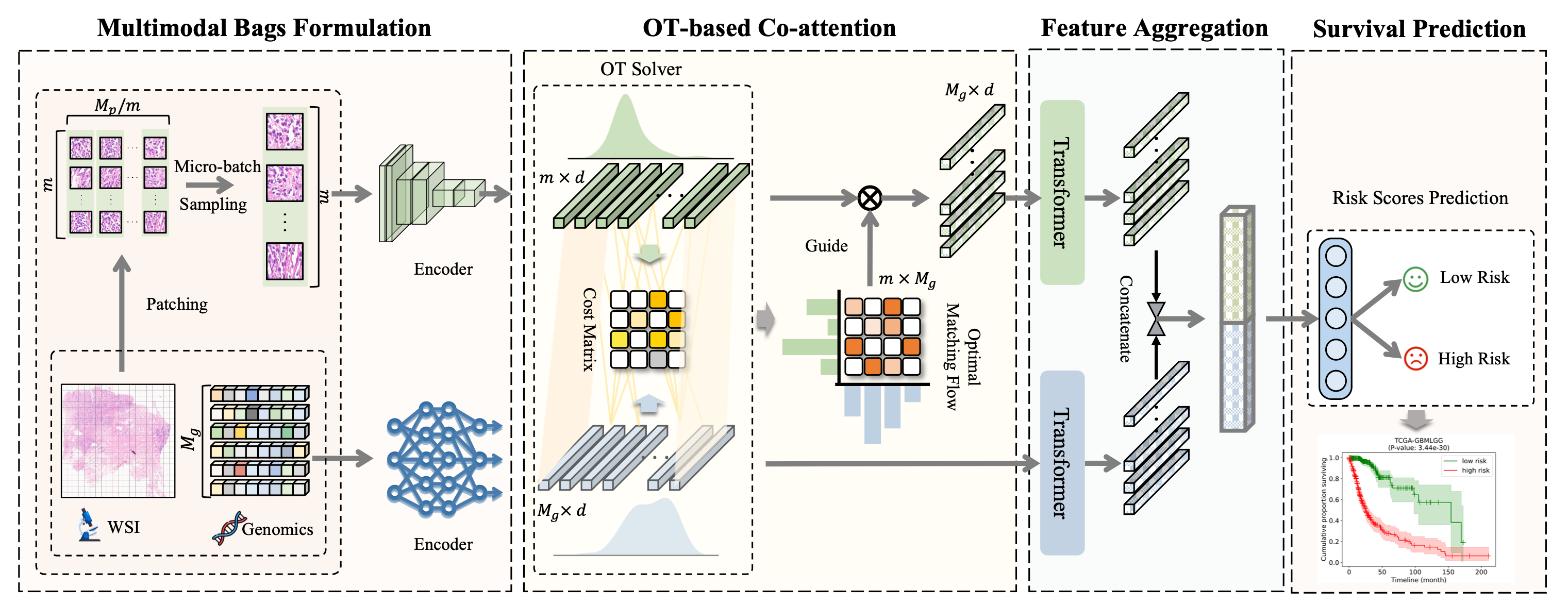
Multimodal Optimal Transport-based Co-Attention Transformer with Global Structure Consistency for Survival Prediction, ICCV 2023.
[arxiv]
[paper]
Yingxue Xu, Hao Chen
@InProceedings{Xu_2023_ICCV,
author = {Xu, Yingxue and Chen, Hao},
title = {Multimodal Optimal Transport-based Co-Attention Transformer with Global Structure Consistency for Survival Prediction},
booktitle = {Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV)},
month = {October},
year = {2023},
pages = {21241-21251}
}Summary: Here is the official implementation of the paper "Multimodal Optimal Transport-based Co-Attention Transformer with Global Structure Consistency for Survival Prediction".
- [09/2024] add an optional argument
--use_micro_batchto enable the Micro-Batch setting. The default value isFalse. If you want to use it, please set it toTrueand specify the batch size for Micro-Batch with--bs_micro. - [04/2024] Upgraded OT module to its GPU version, which allows larger Micro-Batch or the removal of Micro-Batch setting. The Pre-requisites have been updated accordingly. In this case, we have set it to 16384 by default, resulting in notably accelerated training speed.
python==3.9.19
pot==0.9.3
torch==2.2.1
torchvision==0.17.1
scikit-survival==0.22.2- Download diagnostic WSIs from TCGA
- Use the WSI processing tool provided by CLAM to extract resnet-50 pretrained 1024-dim feature for each 256
$\times$ 256 patch (20x), which we then save as.ptfiles for each WSI. So, we get onept_filesfolder storing.ptfiles for all WSIs of one study.
The final structure of datasets should be as following:
DATA_ROOT_DIR/
└──pt_files/
├── slide_1.pt
├── slide_2.pt
└── ...DATA_ROOT_DIR is the base directory of cancer type (e.g. the directory to TCGA_BLCA), which should be passed to the model with the argument --data_root_dir as shown in command.md.
In this work, we directly use the preprocessed genomic data provided by MCAT, stored in folder dataset_csv.
Splits for each cancer type are found in the splits/5foldcv folder, which are randomly partitioned each dataset using 5-fold cross-validation. Each one contains splits_{k}.csv for k = 1 to 5. To compare with MCAT, we follow the same splits as that of MCAT.
To train MOTCat, you can specify the argument in the bash train_motcat.sh stored in scripts and run the command:
sh scripts/train_motcat.shor use the following generic command-line and specify the arguments:
CUDA_VISIBLE_DEVICES=<DEVICE_ID> python main.py \
--data_root_dir <DATA_ROOT_DIR> \
--split_dir <SPLITS_FOR_CANCER_TYPE> \
--model_type motcat \
--use_micro_batch \
--bs_micro 256 \
--ot_impl pot-uot-l2 \
--ot_reg <OT_ENTROPIC_REGULARIZATION> --ot_tau 0.5 \
--which_splits 5foldcv \
--apply_sigCommands for all experiments of MOTCat can be found in the command.md file.
Huge thanks to the authors of following open-source projects:
If you find our work useful in your research, please consider citing our paper at:
@InProceedings{Xu_2023_ICCV,
author = {Xu, Yingxue and Chen, Hao},
title = {Multimodal Optimal Transport-based Co-Attention Transformer with Global Structure Consistency for Survival Prediction},
booktitle = {Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV)},
month = {October},
year = {2023},
pages = {21241-21251}
}This code is available for non-commercial academic purposes. If you have any question, feel free to email Yingxue XU.