Iyar Lin 02 februari, 2024
The xgboost package survival model returns predictions on the hazard
ratio scale (i.e., as $HR = exp(marginal_prediction) in the
proportional hazard function
.
This quantity is equivalent to the type = “risk” in coxph. This package
provides a thin wrapper that enables using the xgboost package to
perform full survival curve estimation.
See also discussion in stackoverflow
Below is a short usage demo.
First, prepare the data:
library(survXgboost)
library(survival)
library(xgboost)
data("lung")
lung <- lung[complete.cases(lung), ] # doesn't handle missing values at the moment
lung$status <- lung$status - 1 # format status variable correctly such that 1 is event/death and 0 is censored/alive
label <- ifelse(lung$status == 1, lung$time, -lung$time)
val_ind <- sample.int(nrow(lung), 0.1 * nrow(lung))
x_train <- as.matrix(lung[-val_ind, !names(lung) %in% c("time", "status")])
x_label <- label[-val_ind]
x_val <- xgb.DMatrix(as.matrix(lung[val_ind, !names(lung) %in% c("time", "status")]),
label = label[val_ind])Below we train an xgboost survival model using the function from the survXgboost package rather than the xgboost package:
# train surv_xgboost
surv_xgboost_model <- xgb.train.surv(
params = list(
objective = "survival:cox",
eval_metric = "cox-nloglik",
eta = 0.05 # larger eta leads to algorithm not converging, resulting in NaN predictions
), data = x_train, label = x_label,
watchlist = list(val2 = x_val),
nrounds = 1000, early_stopping_rounds = 30
)## [1] val2-cox-nloglik:1.986334
## Will train until val2_cox_nloglik hasn't improved in 30 rounds.
##
## [2] val2-cox-nloglik:1.988136
## [3] val2-cox-nloglik:1.999648
## [4] val2-cox-nloglik:2.000725
## [5] val2-cox-nloglik:2.011412
## [6] val2-cox-nloglik:2.010582
## [7] val2-cox-nloglik:2.012655
## [8] val2-cox-nloglik:2.025819
## [9] val2-cox-nloglik:2.027110
## [10] val2-cox-nloglik:2.027684
## [11] val2-cox-nloglik:2.030564
## [12] val2-cox-nloglik:2.034511
## [13] val2-cox-nloglik:2.038002
## [14] val2-cox-nloglik:2.043498
## [15] val2-cox-nloglik:2.052795
## [16] val2-cox-nloglik:2.057096
## [17] val2-cox-nloglik:2.062697
## [18] val2-cox-nloglik:2.074469
## [19] val2-cox-nloglik:2.085677
## [20] val2-cox-nloglik:2.084821
## [21] val2-cox-nloglik:2.097534
## [22] val2-cox-nloglik:2.102217
## [23] val2-cox-nloglik:2.115933
## [24] val2-cox-nloglik:2.123452
## [25] val2-cox-nloglik:2.132746
## [26] val2-cox-nloglik:2.137868
## [27] val2-cox-nloglik:2.136174
## [28] val2-cox-nloglik:2.140923
## [29] val2-cox-nloglik:2.144434
## [30] val2-cox-nloglik:2.149500
## [31] val2-cox-nloglik:2.153794
## Stopping. Best iteration:
## [1] val2-cox-nloglik:1.986334
Next we can predict full survival curves:
# predict survival curves
times <- seq(10, 1000, 50)
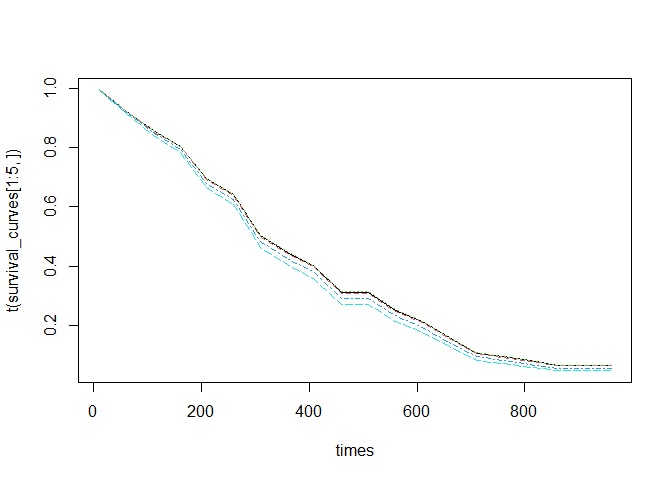
survival_curves <- predict(object = surv_xgboost_model, newdata = x_train, type = "surv", times = times)
matplot(times, t(survival_curves[1:5, ]), type = "l")We can also predict the risk scores like in the original xgboost package:
# predict risk score
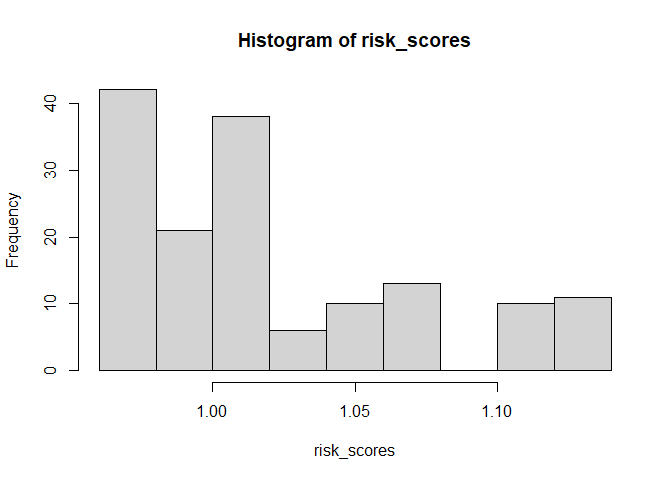
risk_scores <- predict(object = surv_xgboost_model, newdata = x_train, type = "risk")
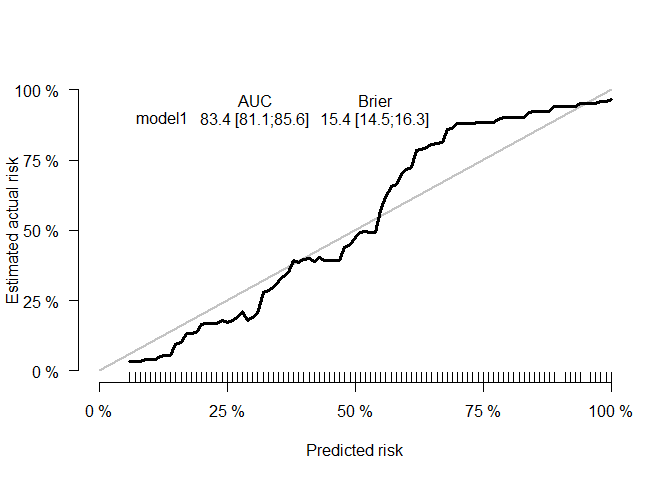
hist(risk_scores)We can see the package can produce survival estimates that are well calibrated
if(require(riskRegression)){
data(cancer, package="survival")
status_mgus2 <- ifelse(mgus2$death == 0, -mgus2$futime, mgus2$futime)
# We use na.pass since XGBoost can predict for missing data
formula_mgus2 <- ~ age + sex + dxyr + hgb + mspike - 1
x_mgus2 <- model.matrix(
formula_mgus2,
model.frame(formula_mgus2, mgus2, na.action = "na.pass")
)
# Note: this model is likely overfitting horribly, but that helps demonstrate calibration
mgus2_model <- xgb.train.surv(
params = list(
objective = "survival:cox",
eval_metric = "cox-nloglik",
eta = 0.2
), data = x_mgus2, label = status_mgus2,
nrounds = 10
)
surv_predictions <- predict(mgus2_model, x_mgus2, type = "surv", times= 60)
score <- Score(list(model1=1-surv_predictions),Surv(futime,death) ~1 ,data=mgus2,
times=60,plots="cal")
plotCalibration(score, rug = TRUE)
}