(Note: -> In Development)
MomentExpansions.jl is an add-on package to DiffEqBiological.jl.
The package facilitates the generation and simulation of ODE moment expansions of chemical reaction networks. For further information, see the following paper: Ale et al, 2013
A simple dimerisation model, comprising two species and two reactions can be defined using the Reaction DSL (domain specific language) syntax described within DiffEqBiological.jl as below:
using MomentExpansions
rn = @reaction_network dimer begin
(k1, k2), X + X ↔ Y
end k1 k2To create a moment expansion of order 2, we can call the approx function, inputting the reaction network and desired truncation order:
expansion = approx(rn, 2)This creates a MomentExpansion container, storing information about the moments, and their ODE equations.
We can create an ODEProblem object just as within any normal DifferentialEquations.jl environment, specifying the time interval, initial species, and parameter values.
# (u0 = [301, 0], the further 3 central moments are assumed to be equal to 0)
u0 = [301, 0.0, 0.0, 0.0, 0.0]
tspan = (0.0, 4.0)
p = [3.32e-3, 0.2]
prob = ODEProblem(expansion, u0, tspan, p)Using the DifferentialEquations library, we can then solve the system.
using DifferentialEquations
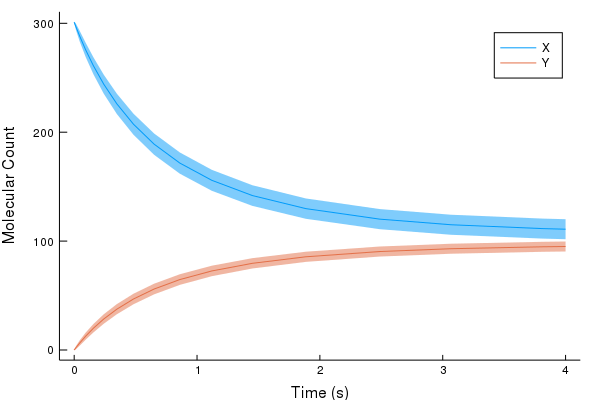
sol = solve(prob, Tsit5())This produces a solution for the means, E[X] and E[Y], and their corresponding second order moments E[(X-E[X])^2], E[(X-E[X])(Y-E[Y])], E[(Y-E[Y])^2].
Using the information about the means, and their second order moments, we can make plots --- e.g. ribbon plots showing the means of X and Y, alongside their respective standard deviations.
using Plots
p1 = plot(sol.t, sol[1,:], ribbon=sqrt.(sol[3,:]), label="X", xlabel="Time (s)", ylabel="Molecular Count", grid=false)
plot!(sol.t, sol[2,:], ribbon=sqrt.(sol[5,:]), label="Y")