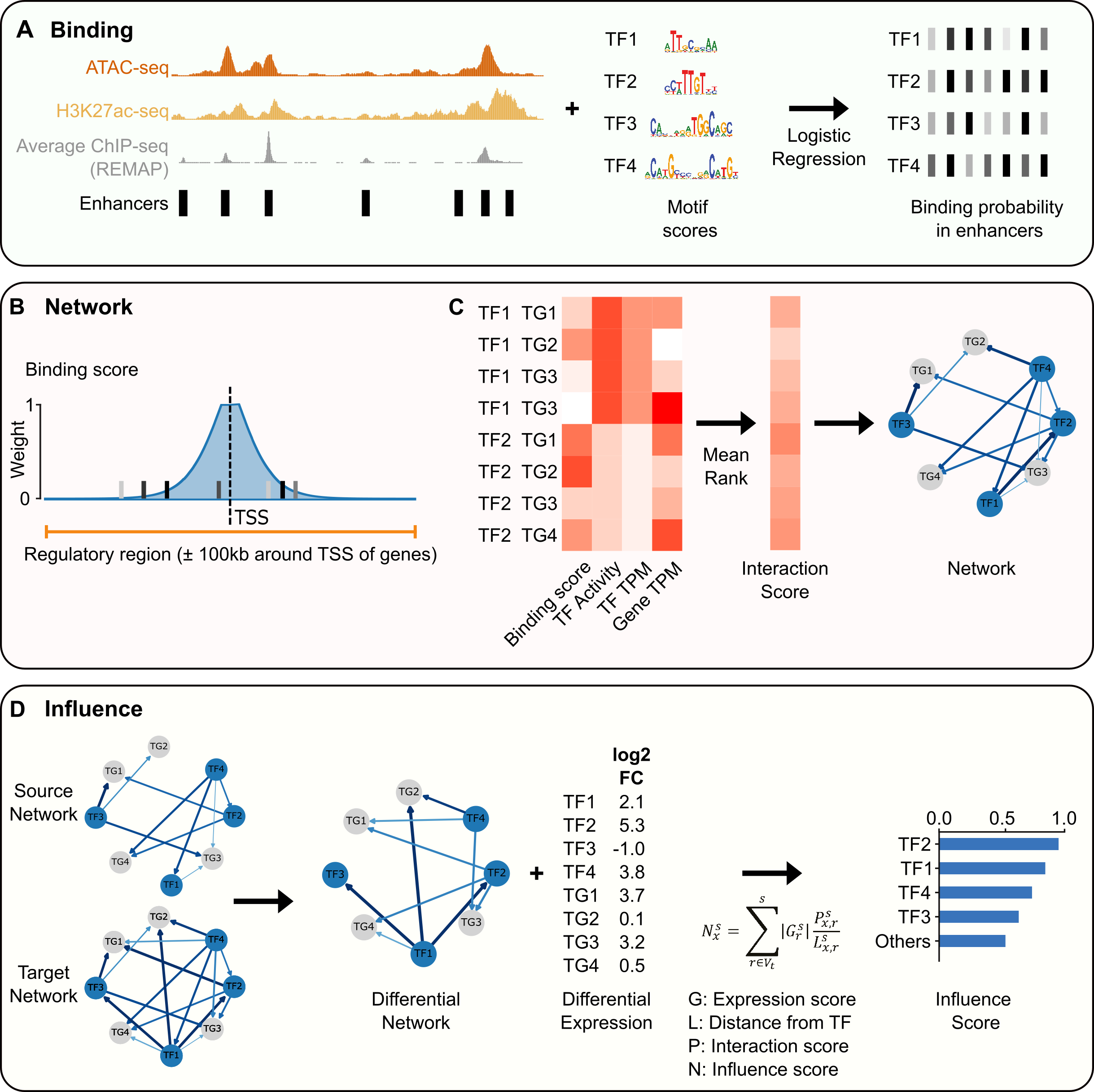
ANANSE is a computational approach to infer enhancer-based gene regulatory networks (GRNs) and to use these GRNs to identify the key transcription factors in cell fate determination. You can use it to generate a shortlist of transcription factors for trans-differentiation experiments, but also to generate cell type-specific gene regulatory networks or to study transcription regulation during development and differentiation. It is written in Python and it contains three command-line scripts: ananse binding, ananse network, and ananse influence. A graphical overview of the tools is shown below.
Read the full ANANSE documentation for detailed installation instructions and usage examples. For documentation on the development version see here.
The most straightforward way to install ANANSE is via conda using the bioconda channel.
1. If you have not used bioconda before, first set up the necessary channels (in this order!). You only have to do this once.
$ conda config --add channels defaults
$ conda config --add channels bioconda
$ conda config --add channels conda-forge
# Create an environment called ananse with all dependencies
$ conda create -n ananse ananse
# Activate the environment
$ conda activate ananse
Don't forget to activate the environment with conda activate ananse whenever you want to use ANANSE.
The latest version, but may not always be stable.
# Activate the environment
$ conda activate ananse
# Install development version
$ pip install git+https://github.com/vanheeringen-lab/ANANSE.git@develop
The three command-line tools (binding, network and influence) can be used separately, but are designed to work together. In general, for a full ANANSE analysis, you would infer binding and calculate the GRN for two (or more) different cell types and then use ananse influence to determine influential TFs for the transition from one cell type to the other.
Before you can use the ANANSE tools, you have to install your genome with corresponding annotation using genomepy. For instance, to use hg38:
genomepy install hg38 --annotation
To predict binding, you need either ATAC-seq and/or H3K27ac ChIP-seq data as BAM files. Using both of these types of data will give the most accurate results, however, either of the two will also work. ANANSE will automatically choose the relevant model depending on which data you use as input. If you have human data, mapped to hg38, you can use a more advanced model based on
ananse binding -A <ATAC.bam> -H <H3k27ac.bam> -o out
To create a gene regulatory network you will need a binding prediction from ananse binding and one or more files with gene expression quantification. The file should have the gene identifier in the first column and a column with TPM as a head. You can use, for instance, the quant.sf from salmon or the abundances.tsv from kallisto, converted to gene-level TPMs with tximport. Here we will run ananse network with 4 threads:
ananse network -b out/binding.h5 -e <gene_tpm.txt> -o network.txt -n 4
To calculate the influence score, you will need two network files from ananse network and a differential expression file. The differential expression file can be generated with DESeq2, where you use the source cell type as the reference. This means that up-regulated genes (log2 fold change > 0) will have a higher expression in the target cell type.
ananse influence -s source.network.txt -t target.network.txt -d source2target.de.tsv -o source2target.out.txt -n 4 -p
- Clone the repo from git.
- Checkout the
developbranch. - Install a development environment with conda:
conda env create -n ananse_dev -f requirements.yaml. - Activate the environment with
conda activate ananse_dev. - Install ANANSE with
python setup.py develop.
ANANSE: an enhancer network-based computational approach for predicting key transcription factors in cell fate determination Quan Xu, Georgios Georgiou, Siebren Frölich, Maarten van der Sande, Gert Jan C Veenstra, Huiqing Zhou, Simon J van Heeringen Nucleic Acids Research, gkab598, https://doi.org/10.1093/nar/gkab598
- The preferred way to get support is through the Github issues page.
- MIT license
- Copyright 2020 © vanheeringen-lab.