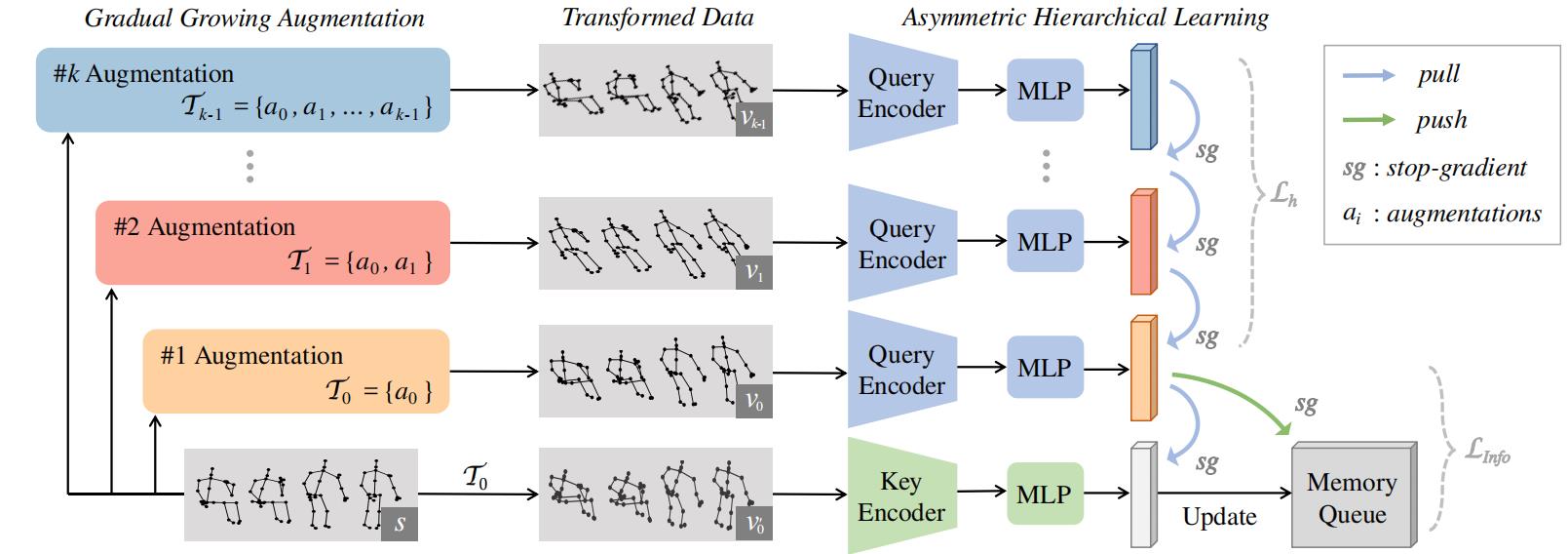
This is an official PyTorch implementation of "Hierarchical Consistent Contrastive Learning for Skeleton-Based Action Recognition with Growing Augmentations" in AAAI 2023.
- Download the raw data of NTU RGB+D and PKU-MMD.
- For NTU RGB+D dataset, preprocess data with
tools/ntu_gendata.py. For PKU-MMD dataset, preprocess data withtools/pku_part1_gendata.py. - Then downsample the data to 50 frames with
feeder/preprocess_ntu.pyandfeeder/preprocess_pku.py.
See the run_cs.sh for the detailed instructions.
You can change the settings in the corresponding .yaml file.
# train on NTU RGB+D xsub joint stream
$ python main.py pretrain_hiclr --config config/release/gcn_ntu60/pretext/pretext_hiclr_xsub_joint.yaml
#linear evaluation on NTU RGB+D xsub joint stream
$ python main.py linear_evaluation --config config/release/gcn_ntu60/linear_eval/linear_eval_hiclr_xsub_joint.yaml
#finetune on NTU RGB+D xsub joint stream
$ python main.py finetune_evaluation --config config/release/gcn_ntu60/finetune/xsub_joint.yamlSimilarly, set the config as the .yaml file in config/transformer_ntu60/ if you want to train a Transformaer-based model.
For three-streams results, we use the code in ensemble_xxx.py to obtain the fusion results. The performance of the released repo is slightly better than that reported in the paper. You can find the pre-trained model weights here (for GCN).
| Model | NTU 60 xsub (%) |
|---|---|
| HiCLR-joint | 77.30 |
| HiCLR-motion | 70.29 |
| HiCLR-bone | 75.59 |
| 3s-HiCLR | 80.94 |
If you find this repository useful, please consider citing our paper:
@article{zhang2022s,
title={Hierarchical Consistent Contrastive Learning for Skeleton-Based Action Recognition with Growing Augmentations},
author={Zhang, Jiahang and Lin, Lilang and Liu, Jiaying},
journal={arXiv preprint arXiv:2211.13466},
year={2022},
}
We sincerely thank the authors for releasing the code of their valuable works. Our code is built based on the following repos.
- The code of our framework is heavily based on AimCLR.
- The code of encoder is based on ST-GCN and DSTA-Net.
This project is licensed under the terms of the MIT license.