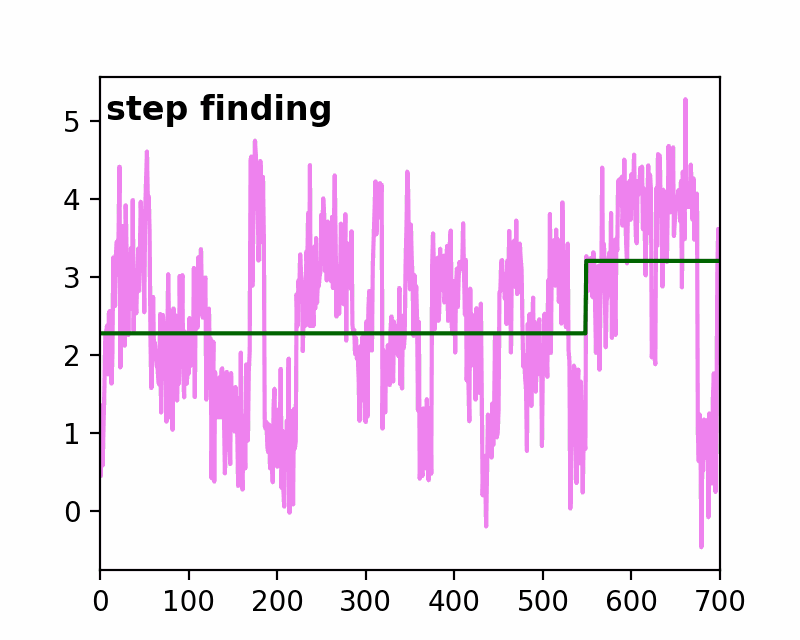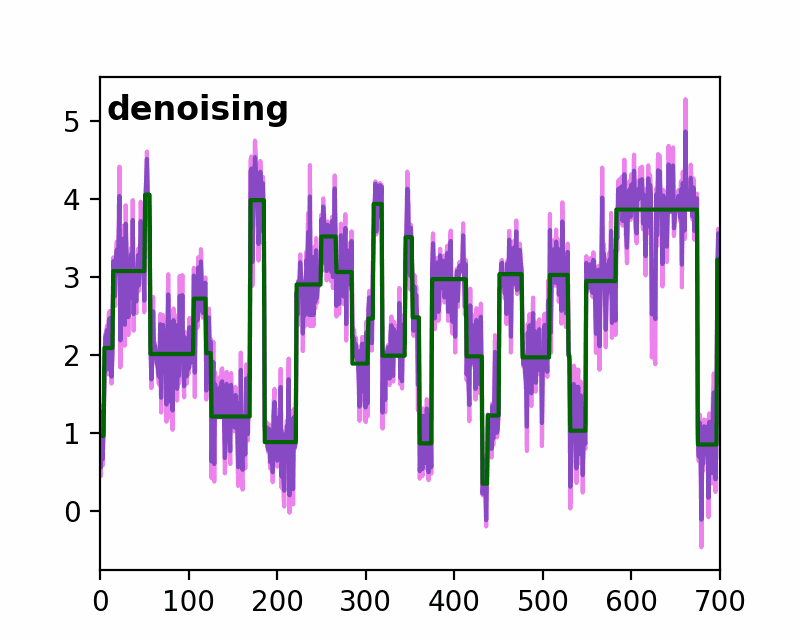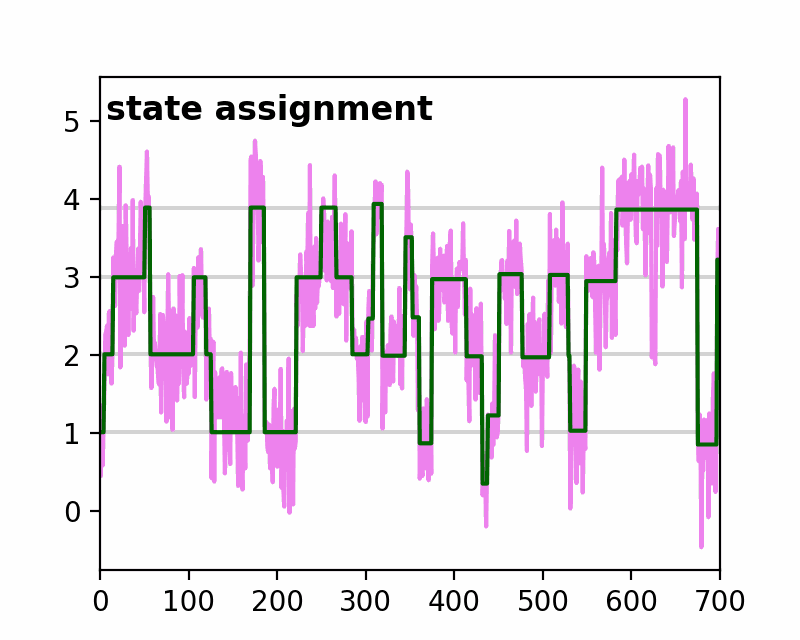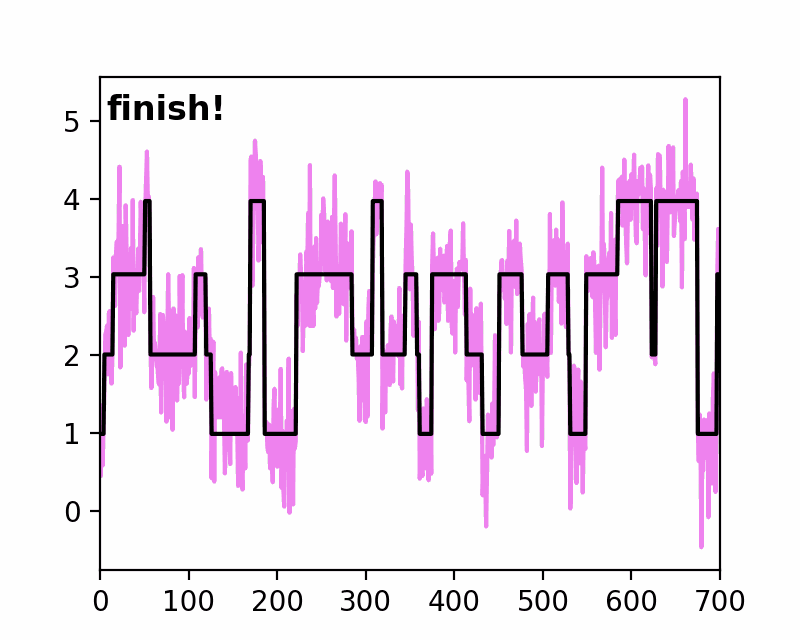
sfHMM (Step-Finding based Hidden Markov Model analysis) provides efficient HMM analysis for varieties of biophysical data. Owing to its accurate parameter estimation workflows, sfHMM can be applied to systems with:
- unknown numbers of states
- high noise
- sparse transition
You can also use sfHMM just for step finding or AIC/BIC based Gaussian mixture model selection. Please refer to "example.ipynb" for basic usages.
- pip installation
Working in a new environment is highly recommended. For example, if you are using Anaconda and Jupyter:
conda create -n myenv python
conda activate myenv
conda install jupyter
then it's ready to install sfHMM.
pip install git+https://github.com/hanjinliu/sfHMM
- cloning from the source
If you want to clone from the source, you need to compile pyx file, or you cannot use sfHMM.motor module. You may need to download C++ build tools for this purpose.
git clone https://github.com/hanjinliu/sfHMM
cd sfHMM
python setup.py build_ext --inplace
I've tested the codes with following versions but may work in older ones.
- python ≥ 3.7, < 3.12
- hmmlearn ≥ 0.2.3
- scikit-learn ≥ 0.24.1
- pandas ≥ 1.1.5
- scipy ≥ 1.6.1 (at least 1.2.0 because
scipy.special.softmaxmust be available) - matplotlib ≥ 3.3.4
This module contains basic classes and functions for sfHMM data analysis.
sfHMM1... sfHMM for single trajectory.sfHMMn... sfHMM for multiple trajectories.hmm_sampling... sample data generation of smFRET-like data.motor_sampling... sample data generation of motor-like data.
This module contains sfHMM classes aiming at analyzing motor-stepping or other sparse transition models.
sfHMM1Motor... sfHMM for single motor-stepping trajectory.sfHMMnMotor... sfHMM for multiple motor-stepping trajectories.
This module contains several step finding algorithms in the same API. My efficient implementation enables analysis of 100,000 data points within ~1 sec!
GaussStep... step finding for Gauss distribution [1].PoissonStep... step finding for Poisson distribution.SDFixedGaussStep... step finding for Gauss distribution with fixed standard deviation.TtestStep... step finding using T-test [2].BayesianPoissonStep... step finding in Bayesian method [3].
This module contains classes that inherit sklearn.mixture.GaussianMixture while making Gaussian mixture model clustering much easier.
GMMs... Fitting Gaussian mixtures and model selection.
This module contains input/output functions that are suitable for sfHMM data analysis or other one-dimensional signal processing.
read... Not recommended now. Use "read" method in sfHMM classes insteadread_excel... Load all the sheets from Excel files.save... Not recommended now. Use "save" method in sfHMM classes instead
This module contains interactive multi-channel viewer for 1-D data visualization (work in progress).
TrajectoryViewer... Qt viewer object.
All the parameters are optional.
sg0... The parameter used in denoising process.psf... The parameter used in step finding.krange... Range of the number of hidden states to test.model... Distribution of signal. Gaussian and Poissonian are supported now.name... Name of the object.
run_all()... Run all the needed algorithm in the most plausible condition.step_finding()... Step finding by likelihood maximization.denoising()... The standard deviation of noise is cut off tosg0.gmmfit()... Gaussian mixture model clustering.hmmfit()... HMM parameter initialization and optimization.plot()... Visualize the results of sfHMM analysis.tdp()... Show the results in pseudo transition density plot.read()... Load such as csv, txt, dat files, or the first sheet of Excel files.save()... Save sfHMM analysis results.view_in_qt()... Open aTrajectoryViewerand interactively view the trajectories.
If you found sfHMM useful, please consider citing our paper.
A fast and objective hidden Markov modeling for accurate analysis of biophysical data with numerous states
Hanjin Liu, Tomohiro Shima
bioRxiv 2021.05.30.446337; doi: https://doi.org/10.1101/2021.05.30.446337
[1] Kalafut, B., & Visscher, K. (2008). An objective, model-independent method for detection of non-uniform steps in noisy signals. Computer Physics Communications, 179(10), 716-723.
[2] Shuang, B., Cooper, D., Taylor, J. N., Kisley, L., Chen, J., Wang, W., ... & Landes, C. F. (2014). Fast step transition and state identification (STaSI) for discrete single-molecule data analysis. The journal of physical chemistry letters, 5(18), 3157-3161.
[3] Ensign, D. L., & Pande, V. S. (2010). Bayesian detection of intensity changes in single molecule and molecular dynamics trajectories. Journal of Physical Chemistry B, 114(1), 280–292.