Scientists spend hours reading the most recent scientific literature to gain state of the art knowledge of gene to disease relationships. To help scientists discover relevant literature faster, GeDi will be developed to analyze new scientific literature at the instant of publication and extract gene to disease relationships that co-occur in that publication. GeDi then displays the gene to disease relationships of scientific interest to scientists along with a link back to the original publication. There are similar applications available commercially, but GeDi gives scientists a competitive advantage by providing up to the minute and personalized updates of newly discovered gene to disease relationships.
-
System must allow end users via a UI application to search for a gene and find all related diseases we have found from scientific literature.
-
System must provide an endpoint that allows for passing the URL location of a scientific document for the system to collect, parse, annotate, and make available to the search application.
-
An NLP or other text-mining analysis process must be executed on collected scientific literature to extract gene to disease relationships.
-
System must store all annotations found in a scientific document, along with associated metadata (article title, link back to article, etc)
-
System must be well tested with comprehensive unit and integration test suites.
-
Production environment must be suitable for future scaling needs.
-
Continuous delivery must be supported. Full test suites must be executed successfully before all branch merges. Successful merging into the main branch deploys the application to production.
-
The analysis application consumes large resources, so the system must provide production monitoring of the analysis tier so that capacity and cost can be closely monitored.
-
Callers of the analysis application shouldn't have to wait for analysis to complete to get a result from the analysis api. The API should respond instancty and process articles in the background asynchronously.
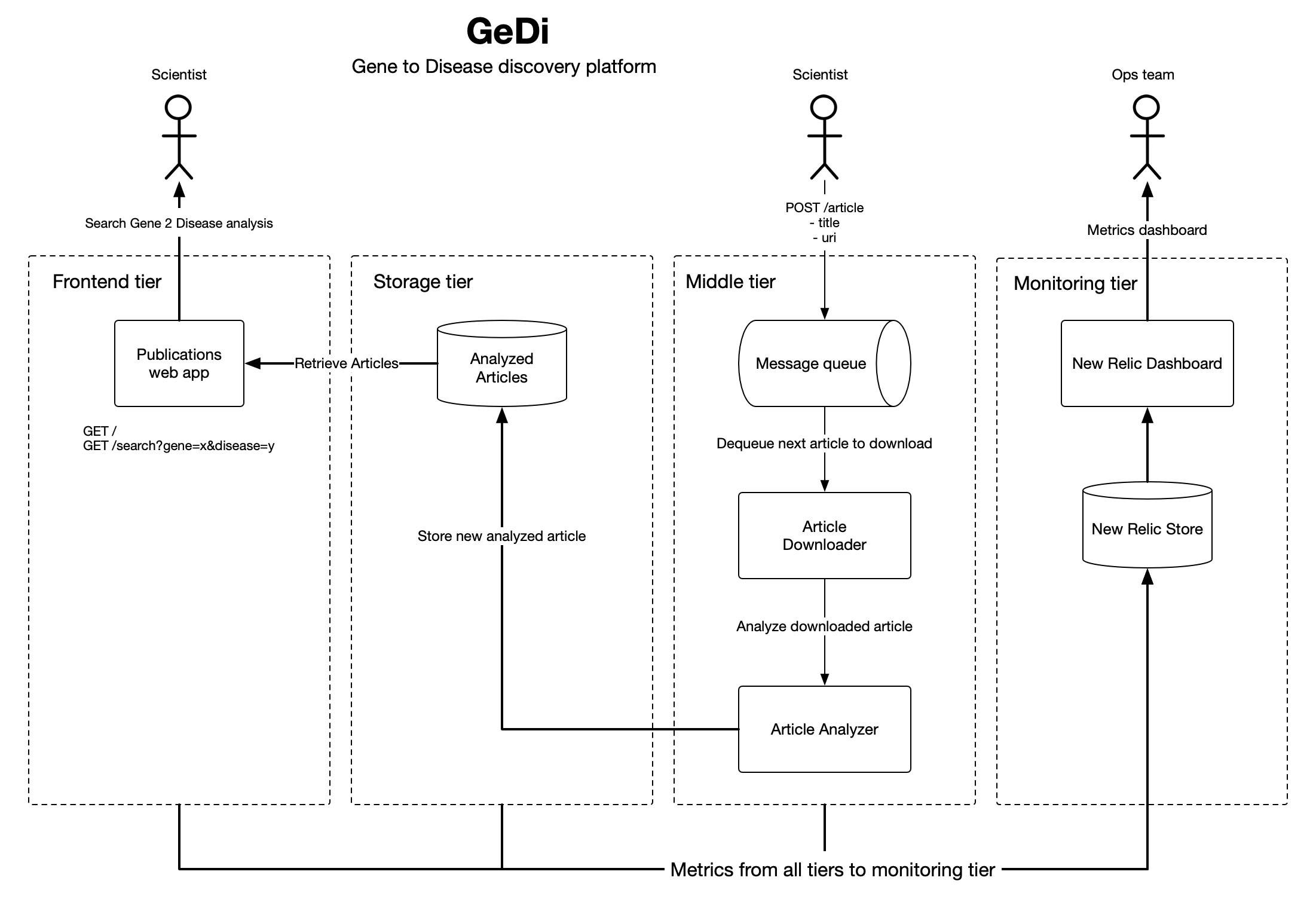
The application's logical architecture is split into 4 tiers, front-end, storage, middle, and monitoring. The front-end application serves article analysis served from the storage layer. The middle tier handles all the business logic and long running processes of the application including downloading and analyzing articles. The monitoring layer gives us a window into our application metrics to monitor usage and performance.
I chose Python as the programming language because of the rich machine learning libraries available. For the NLP analyzer, I chose SciSpacy for it's robust medical named entity recognition features that were perfect for analyzing biology articles.
There were a lot of requirements to cover in a short period of time, so I chose Flask as the the web application framework for it's flexibility, speed of development, and testing tools.
For persistence, I chose Postgresql because I needed a relational data store and Postgres is very well supported and scalable.
I chose RabbitMQ for messaging because while other message queues like Kafka are good for very high throughput applications, a lightweight system like Rabbit would fit the requirements nicely for long running processes like text analysis.
Below is the documentation outlining how I implemented each part of the grading rubric.
Users interact with GeDI data through a simple search UI. The web application's only responsibility is to respond to user search requests. I implemented a Flask app to provide a sufficient web framework for this application. All business logic is handled in the middle tier, and persistance and data integrity is handled in the storage tier. I've used Heroku for hosting.
The deployed web application can be found here
The web application code can be found here
The data collector is bundled with the data analysis application, and can be found here. The data collection app downloads and then parses the pdf into plaintext before passing to the SciSpacy analyzer. Downloading, parsing, and analyzing a pdf is a long running task, so we use event collaboration messaging to process the job in the background using Celery and RabbitMQ. More on the even collaboration messaging implementation in the following secitons.
After downloading and parsing a pdf, we pass the PDF contents along with the newly created article_id over to the SpacyAnalyzer. The SpacyAnalyzer executes the en_ner_bionlp13cg_md model over the parsed pdf, and extracts the gene to disease relationships. Extracted gene to disease relationships are then saved in Postgres along with the article_id for future reference.
There is a comprehensive suite of unit tests built using the Pytest framework. Suite of tests for the webapp can be found here, and the suite of tests for the analyzer/downloader/parser can be found here.
I found that the user requrements were very testable, and I was able to produce tests for the web ui, data downloader, analyzer, database, and NLP processes.
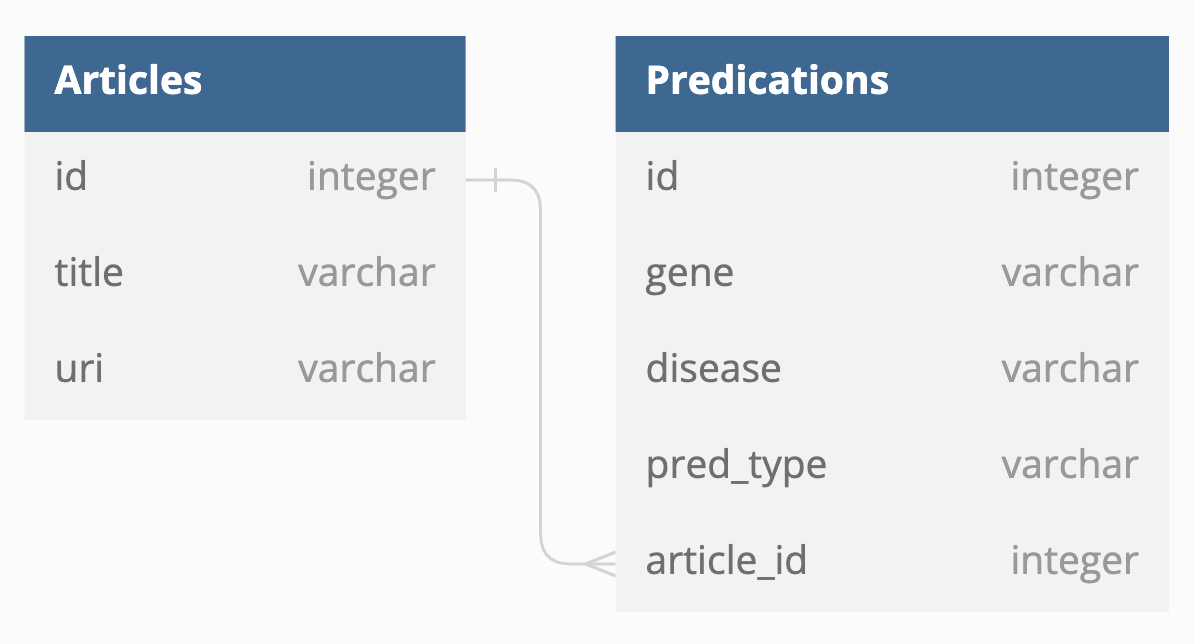
This is the tier where references to literature, associated metadata (title, uri etc), and annotations we extract from the literature is persisted and then used by the web application for retrieval. PostgreSql will be sufficient for our scaling needs, and is supported on all cloud providers including Heroku. I've implemented object relational mapping using SQLAlchemy library and database migrations using the Alembic library.
Stores each article that was downloaded and analyzed by the analyzer application.
- title: Title of the analyzed article
- uri: Location of the analyzed article
Stores each predication found and links the NLP discovery back to the article.
- gene: Left side of the extracted relationship. Left side will always be a gene.
- disease: Right side of the extracted relationship. Right side will always be a type of cancer.
- pred_type: The relationship type. Co-occurrance means that the detected gene and disease appear in the same sentence.
- article_id: Foreign key for the Article the predication was found in.
A POST API has been set up using the Flask Restful framework to allow submission of articles to be downloaded, parsed, and analyzed. POST API can be found here
Heroku is used to host the production environment. Heroku provides us a platform for scalable storage, compute, messaging as well as storing application secrets, environment variables, and continuous deployment. The URL for the production web application can be found here.
For the webapp, there is an integration test that tests integration of the data store, the search page, and the resuts page. Testing of the search submission page to ensure the gene "TP53" is present, and follows up with a check on the search results page for the expected results from the data store. Integration test can be found here.
I used the MagicMock framework to mock the downloader's network call for fast executing unit tests and to remove the network service requirement for the tests. The mock object implementation can be found here
I used Github actions to fulfill the continuous integration requirement. I set up a branching workflow with a single main branch that can only be merged into via pull request, and then only when all of the unit and integration tests pass for the respective apps. CI configuration for the gedi webapp can be found here, and the CI configuration for the gedi analyzer app can be found here.
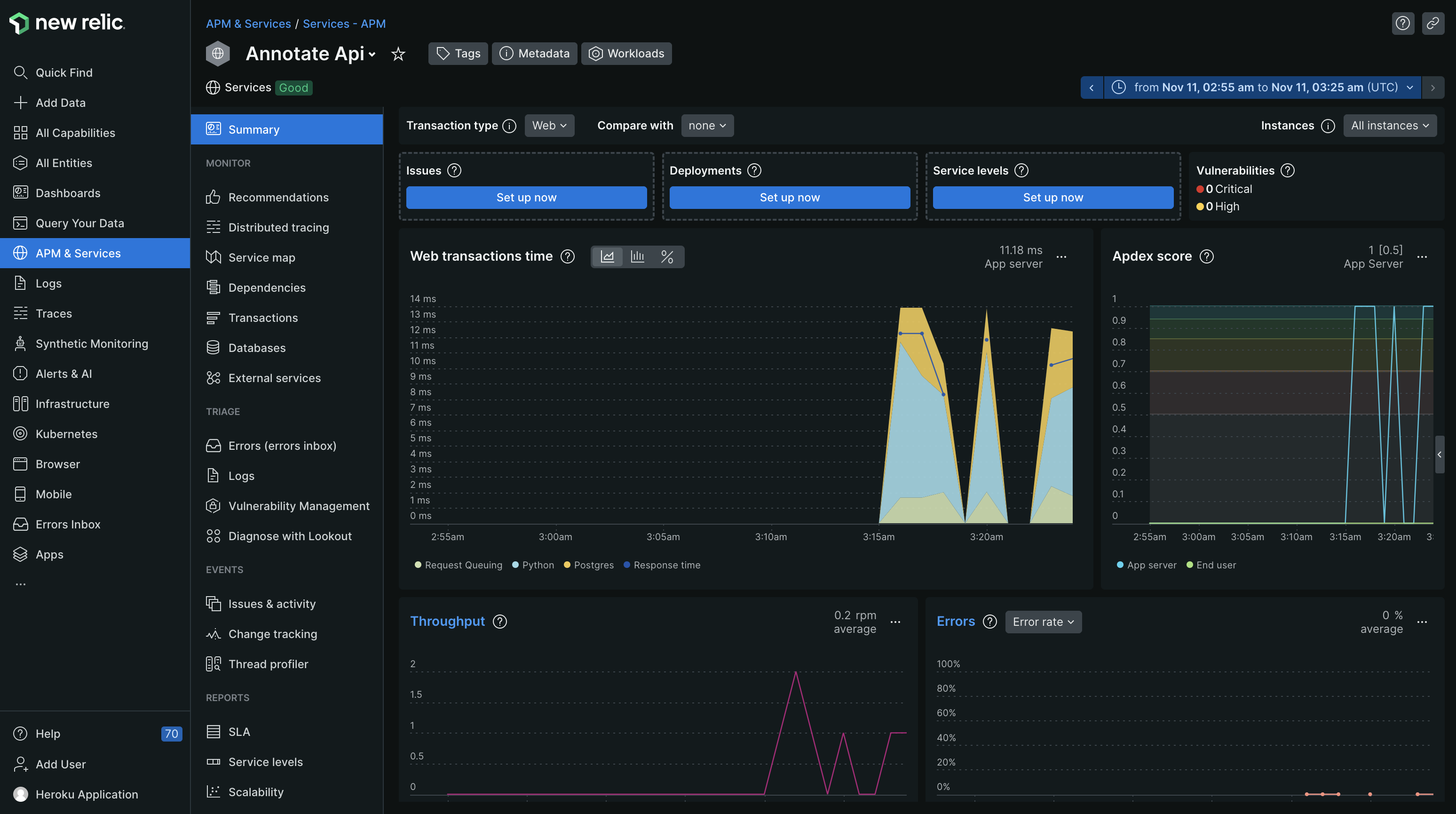
New Relic was used for production monitoring. After activating the New Relic service on Heroku, I installed the New Relic Python library, and activated the New Relic agent in my Procfile found here. New Relic allows for monitoring requests, throughput, errors, CPU/memory usage, and other useful metrics.
This was the most fun part of building the application. The analyzer application can take up to 30 seconds to process and analyze a large document. To improve response times, I implemented a background worker using Celery and RabbitMq. Celery works very well with RabbitMq, and both are supported on the Heroku platform as a persistent worker and add-on respectively. When an article is submitted via the POST api, the API immediately responds with an article id that the caller can use to poll and check if the job has completed. At the same time I submit the job into the Rabbit queue using the Celery .delay() method to be picked up by the next available Celery worker and then processed. According to the New Relic instrumentation, it takes on average 14 seconds to process an an article.
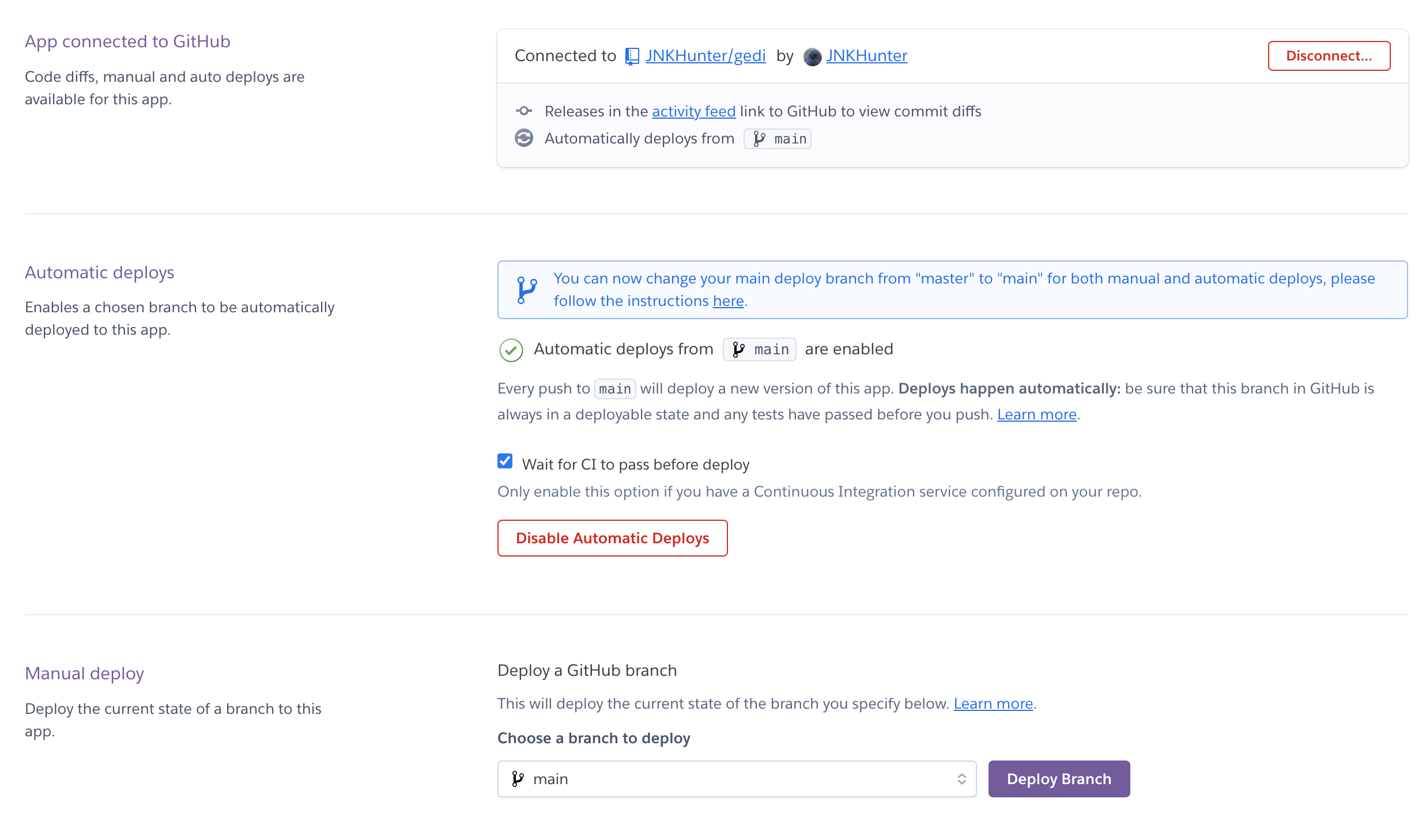
Continuous delivery is achieved by connecting the main branches of each application with a Heroku application. When all tests pass and code is integrated into the main branch, Github signals to Heroku that the main branch was updated, and a process I set up in Heroku deploys the newly built app into the production environment. Screenshot of the CD configuration can be seen below.
git clone git@github.com:JNKHunter/gedi.git
then
git clone git@github.com:JNKHunter/gedi_analyzer.git
Set the following environment variables:
export DATABASE_URL=sqlite:////path/to/your/dev/db/location.db
export MESSAGE_BROKER=amqp://guest@localhost//
With docker/docker compose installed, start up the docker containers in a new window from the project root directory. This command will start up a postgres server locally for testing, psqladmin, and RabbitMQ
docker compose up
The gedi project uses Python 3.11.5 The gedi_analyzer uses Python 3.9.18
Create two separate conda or virtualenv environments for each project using the corresponding Python versions. Any Python commands in these instructions that are run from either the gedi or gedi_analyzer application directories are assumed to be run in those virtualenv or conda environments.
After creating the environments, install the requred packages for each project using requrements.txt in the respective projects.
To migrate the database, cd into the gedi directory and run the migration script
cd gedi
In a separate terminal window in the project root directory
flask db migrate
From the gedi directory
pytest
Form the gedi_analyzer directory
python -m unittest discover -s tests -p "test_*.py"
From the gedi directory
flask run
From the gedi_analyzer directory
flask --app main run -p 5001
From the gedi_analyzer directory
celery -A tasks worker --loglevel=INFO