Co-Training for Crystal Synthesizability Prediction
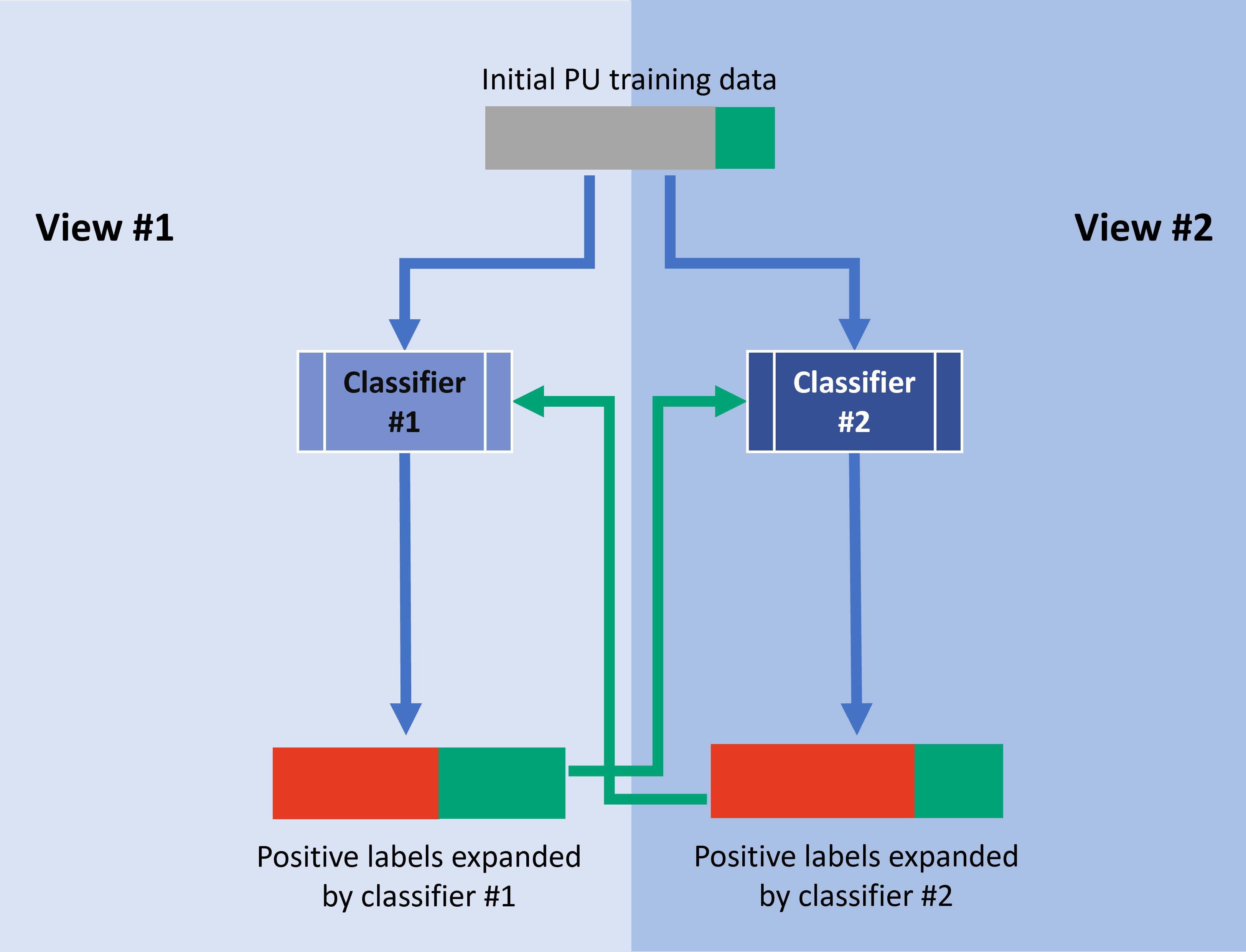
SynthCoTrain is a materials-informatics package which predicts the synthesizability of crystals. The nature of the problem is a semi-supervised classification, in which we access only to positively labeled and unlabeled data points. SynCoTrain does this classification task by combining two semi-supervised classification methods: Positive and Unlabeled (PU) Learning and Co-training. The classifiers used in this package are ALIGNN and SchNetPack.
The final model achieves a notable true-positive rate of 96% for the experimentally synthesized test-set and predicts that 29% of the theoretical crystals are synthesizable. These results go beyond the scope of thermodynamic stability analysis alone. This work carries significant implications, including the filtration of structural predictions from high-throughput simulations to identify synthesizable candidates.
It is recommended to create a virtual environment with mamba and miniforge to install the different packages easily. Start by installing mamba according to the instructions here.
Then we can create our environment and activate it. Let's call it sync:
mamba create -n sync python=3.10
mamba activate syncThe easiest way of installing all the required libraries is to take advantage of the requirements.txt file:
pip install -r requirements.txtThen, you can clone this repository in your preferred path and install it with the following commands:
cd /path/to/parent/directory
git clone https://github.com/sasanamari/SynCoTrain.git
cd SynCoTrain
pip install .If the first method does not conclude successfully, you can try installing the required libraries manually. Start by installing the ALIGGN model by following these instructions or executing the commands below line by line:
mamba install pytorch torchvision torchaudio pytorch-cuda=11.8 -c pytorch -c nvidia
mamba install -c dglteam/label/cu118 dgl
pip install alignn
pip install dgl==1.0.1+cu117 -f https://data.dgl.ai/wheels/cu117/repo.htmlNext, you'll need to install SchNetPack. To ensure compatibility with SynCoTrain, you could use the exact version of SchNetPack that was used during development. Follow the commands below to clone the SchNetPack repository, navigate to it, checkout the specific commit, and install:
git clone https://github.com/atomistic-machine-learning/schnetpack.git
cd schnetpack
git checkout 6fe78ef23313ef1c7c07991cd046ca4d49da8717
pip install .The final required package is Pymatgen which can be installed according to the instruction here or simply using command:
mamba install --channel conda-forge pymatgenAfter installing all the required packages, SynCoTrain can be installed as explained in method #1 by cloning the repository in an appropriate path:
cd /path/to/parent/directory
git clone https://github.com/sasanamari/SynCoTrain.git
cd SynCoTrain
pip install .You don't need to train the model from scratch if you are only interested in predicting synthesizability. The current version of SynCoTrain has been trained to predict the synthesizability of oxide crystals.
To this end, you may use the checkpoint file predict_target/synth_final_preds/checkpoint_120.pt and follow the instructions on ALIGNN repository on how to use a pretrained model.
Alternatively, you can deposite the POSCAR files of the crystals of your interest in a directory in predict_target/label_alignn_format/poscars_for_synth_prediction/<your_directory_name>. The command below predicts the synthesizability of these crystals and saves them in synth_pred.csv in the same directory:
python predict_target/synthesizability_predictor.py --directory_name <your_directory_name>The results will be saved in predict_target/label_alignn_format/synth_preds.csv.
This package provides two auxiliary experiments to evaluate the model further. The first one includes running the regular experiments on only 5% of the available data. This is useful for checking the workflow of the code, without waiting for weeks for the computation to conclude. Please note that quality of results will suffer, as there is less data available for training. The second auxiliary experiment consists of classifying the stability of crystals based on their energy above hull, through the same PU Learning and Co-training code. The utility of this experiment is that, unlike a real case of Positive and Unlabeled Learning, we have access to correct labels of the unlabeled class. As stability is highly related to synthesizability, the quality of this experiment can be used as a proxy to judge the quality of the main experiment. We are mainly interested to see whether the real true-positive-rate of these experiments are close in value to the true-positive-rate produced by PU Learning.
To replicate the results of this library, you need to run the scripts made for running each PU experiment. There are three experiments for each of the base classifiers, with pre-defined data handling. Each experimment consists of 60 iterations of PU learning.
First, the base experiment is run with each model. Next, each model can be trained on the additional psuedo-labels provided by the other model.
Please note that these experiments are rather long. Using a NVIDIA A100 80GB PCIe GPU, each experiment took an average of one week or more to conclude. So, for the full-data experiment, you may want to use the nohup command, as shown later.
It is recommended not to run simultanous experiments on the same gpu, since you run the risk of overflowing the gpu memory and crashing the experiment mid-way.
Before co-training, we need to train our models separately on our PU data; we call this step iteration "0". The code for running the SchNetPack part of this step could be:
mamba activate sync
python pu_data_selection.py --experiment schnet0
nohup python pu_schnet/schnet_pu_learning.py --experiment schnet0 --gpu_id 0 > nohups/schnet0_synth_gpu0.log &In case you have access to multiple GPUs, the --gpu_id parameter can be changed accordingly. Similarly for the ALIGNN experiment we have:
mamba activate sync
python pu_data_selection.py --experiment alignn0
nohup python pu_schnet/schnet_pu_learning.py --experiment alignn0 --gpu_id 0 > nohups/alignn0_synth_gpu0.log &After each experiment is concluded, the data needs to be analyzed to produce the relevant labels for the next step of co-training. The code for the analysis of results of SchNetPack is
python pu_schnet/schnet_pu_analysis.py --experiment schnet0
and for ALIGNN:
python pu_alignn/alignn_pu_analysis.py --experiment alignn0
From this point, it matters that the experiments are executed in their proper order. Before each PU experiment, the relevant data selection needs to be performed. After each PU experiment, the analysis of the results are needed to produce the labels for the next iteration. The commands to run these experiments can be found on synth_commands.txt.
The correct order of running the experiments starting from alignn0 is: alignn0 > coSchnet1 > coAlignn2 > coSchnet3 and for the other view, starting from schnet0: schnet0 > coAlignn1 > coSchnet2 > coAlignn3
The auxiliary stability experiments can be run with almost the same commands, except for an extra --ehull015 True flag. The relavant commands are stored in stability_commands.txt.
After the final round of predictions, the predictions are averaged to produce the final labels. Then, the final ALIGNN-based model is trained as follows:
python predict_target/label_by_average.py
python predict_target/preper_alignn_labels.py
nohup python predict_target/train_folder.py > nohups/synth_predictor.log &