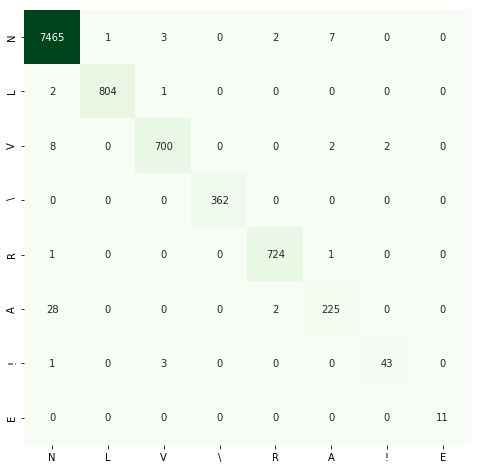
The repository contains code for Master's degree dissertation - Diagnosis of Diseases by ECG Using Convolutional Neural Networks. Only CNN neural network models are considered in the paper and the repository. As a part of the work, more than 30 experiments have been run. The table with all experiments and their metrics is available by the link
The best 1D and 2D CNN models are presented in the repository The repository follows config principle and can be run in the following modes:
- Training - use
train.py --config configs/training/<config>.jsonto train the model - Validation - use
inference.py --config configs/inference/config.jsonto validate the model - Pipeline - use
pipeline.py --config configs/pipelines/config/jsonto test the model using ECG data (i.e. data generation, running, visualization the results)
All available models and all necessary information are described below
Python 3.7 and PyTorch are used in the project GitHub actions are used for installing dependencies and training implemented models
Program - Data Mining Department - Computer Science
Principal Investigator - Nikolai Yu. Zolotykh National Research University - Higher School of Economics
- Cardiologist-Level Arrhythmia Detection with Convolutional Neural Networks
- ECG Heartbeat Classification Using Convolutional Neural Networks
- Electrocardiogram Generation and Feature Extraction Using a Variational Autoencoder (encoder only)
- Author's EcgResNet34
- ECG arrhythmia classification using a 2-D convolutional neural network
- MobileNetV2
- EfficientNetB4
| name | type | model | accuracy | val loss |
|---|---|---|---|---|
| exp-025 | 1D (1x128) - [PEAK[t] - 64, PEAK[t] + 64] | https://arxiv.org/pdf/1707.01836.pdf | 0,9827 | 0,0726 |
| exp-030 | 1D (1x128) - [PEAK[t] - 64, PEAK[t] + 64] | https://ieeexplore.ieee.org/stamp/stamp.jsp?arnumber=8952723 | 0,9864 | 1,5 |
| exp-031 | 1D (1x128) - [PEAK[t] - 64, PEAK[t] + 64] | https://arxiv.org/pdf/2002.00254.pdf | 0,9886 | 0,15 |
| exp-018 | 2D (128x128) - [PEAK[t] - 64, PEAK[t] + 64] | https://arxiv.org/pdf/1804.06812.pdf | 0,9920 | 0,1 |
| exp-013 | 2D (128x128) - [PEAK[t] - 64, PEAK[t] + 64] | MobileNetV2 | 0,9934 | 0,088 |
| exp-021 | 2D (128x128) - [PEAK[t-1] + 20, PEAK[t+1] - 20] | EfficientNetB4 | 0,9935 | 0,062 |
| exp-029 | 1D (1x128) - [PEAK[t] - 64, PEAK[t] + 64] | Novel EcgResNet34 | 0,9938 | 0,0500 |
Training quick start:
- Download
and unzip files into
mit-bihdirectory - Install requirements via
pip install -r requirements.txt - Generate 1D and 2D data files running
cd scripts && python dataset-generation-pool.py - Create
jsonannotation files- For 1D model -
cd scripts && python annotation-generation-1d.py - For 2D model -
cd scripts && python annotation-generation-2d.py
- For 1D model -
- Run training -
python train.py --config configs/training/<config>.json
See CI examples for each model
Using EcgResNet34 model as it shows the best metrics
- Install requirements via pip install -r requirements.txt
- Create directory named
experiments - Download the archive and unzip its content into
experimentsdirectory - Download WFDB format data
- Change
ecg_datapath inconfigs/pipelines/config.jsonwith no extension
{
...
"ecg_data": "./mit-bih/100",
...
}
- Run pipeline -
python pipeline.py --config configs/pipelines/config.json
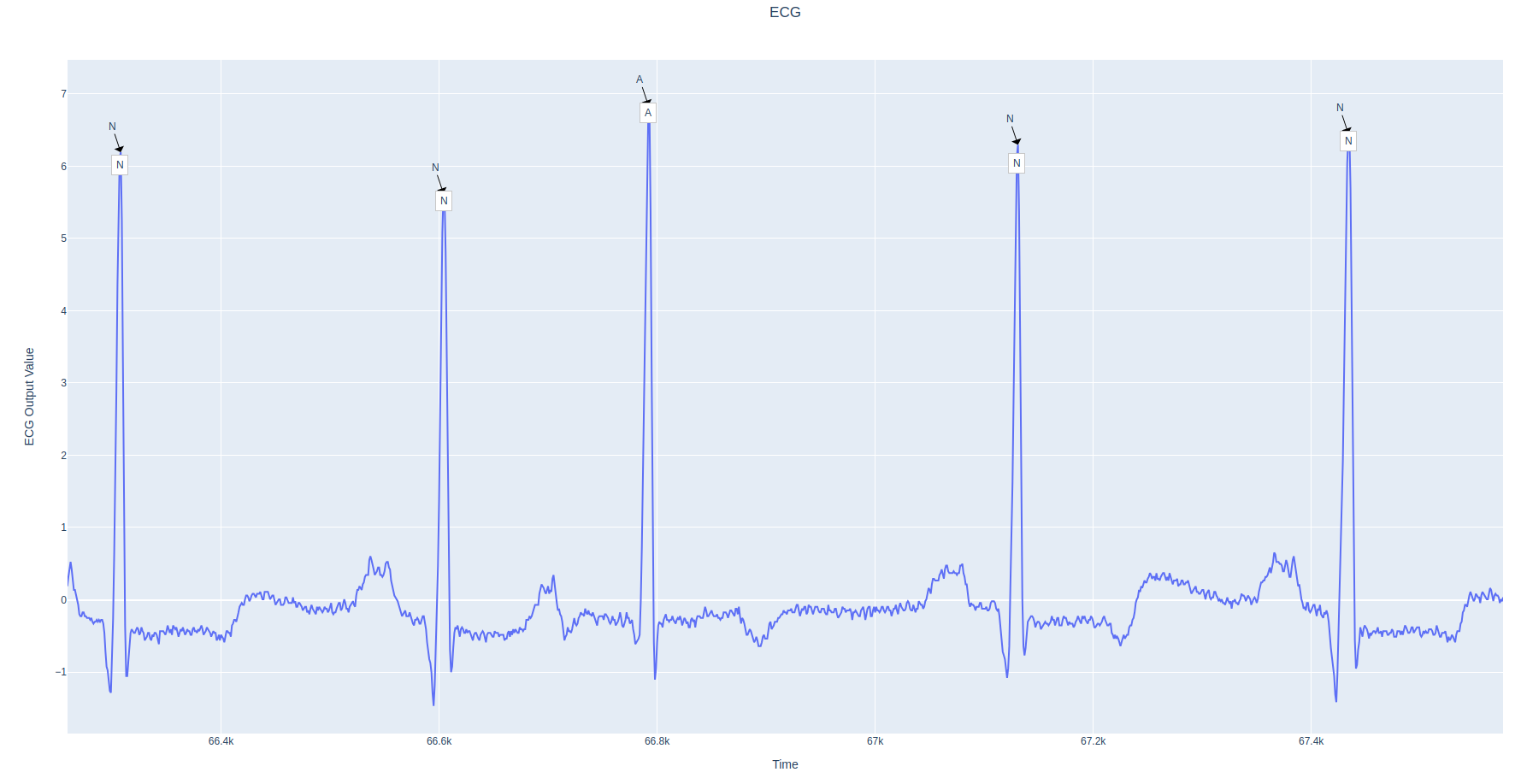
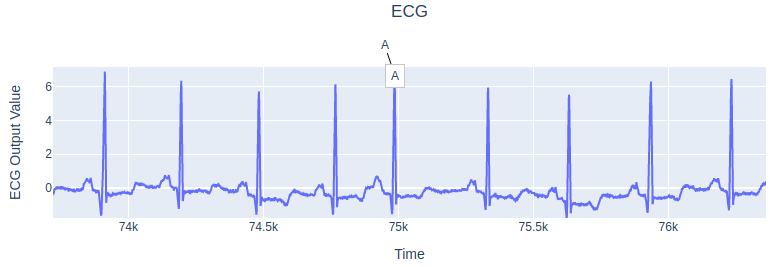
The results will be saved as HTML file in experiments/EcgResNet34/results directory
The code of all experiments described in the table is in branches experiments/exp-XXX
The repository contains Jupyter Notebooks (see notebooks folder)
Please give a ⭐️ if this project helped you
This project is licensed under the MIT License